In these notes, I’ll discuss the PET-UNK-abc197b8-1 design (a dihydrouracil) as a means to merge 3-aminopyridine-like and quinolone series. Some supporting information for the design is provided as discussion for the original submission. The design should be considered as being derived from the VLA-UCB-50c39ae8-3 design which had been submitted previously. When submitting the PET-UNK-abc197b8-1 design, I was actually unaware that it (and VLA-UCB-50c39ae8-3) could effectively merge the 3-aminopyridine-like and quinolone series.
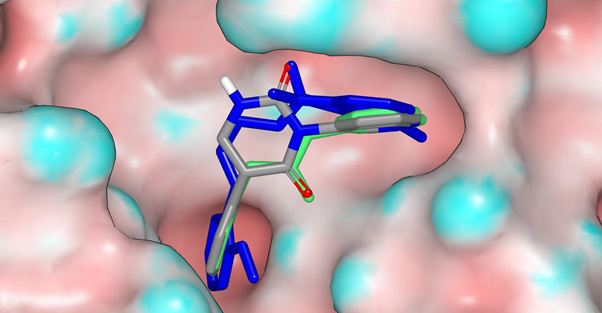
ADA-UCB-6c2cb422-1 is shown below with the molecular surface of the protein colored by curvature (X10959). The amide carbonyl oxygen accepts a hydrogen bond from the backbone NH of E166 and the region around this hydrogen bond donor region of the molecular surface is relatively flat and solvent-exposed.
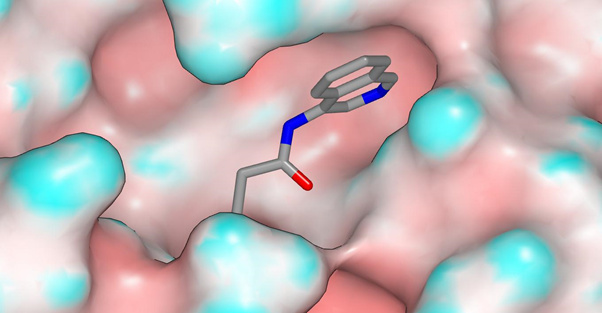
EDJ-MED-6af13d92-3 is shown below with the molecular surface of the protein colored by curvature (X11294). The amide carbonyl oxygen appears to accept hydrogen bonds from the backbone NH of G143 and the sidechain NH2 of N142. This region of the protein molecular surface of the protein in contact with the amide carbonyl oxygen of EDJ-MED-6af13d92-3 is less solvent-exposed than the region of the X10959 molecular surface that makes contact with the amide carbonyl of ADA-UCB-6c2cb422-1.
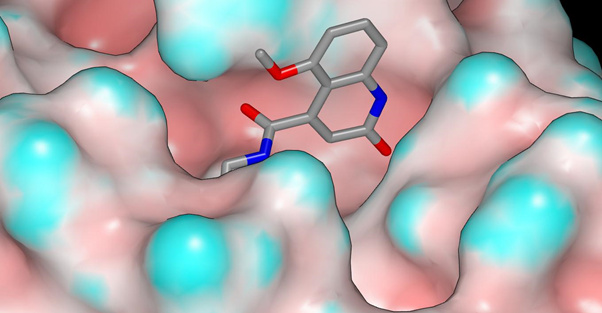
A potential binding mode for PET-UNK-abc197b8-1 was generated using Hybrid from OpenEye and the X10959 crystal structure. This is shown below with the crystallographic ligands ADA-UCB-6c2cb422-1 (green) and EDJ-MED-6af13d92-3 (blue). The molecular surface is for the X11294 crystal structure and has been colored by curvature. The structures in this overlay can be downloaded in pdb format Mpro-x11294_0A_with_ligands.pdb (247.2 KB).
The amidic NH in the dihydrouracil could serve as a vector for structural elaboration. Replacing this NH with CH2 would be expected to weaken the carbonyl oxygen atoms as hydrogen bond acceptors.
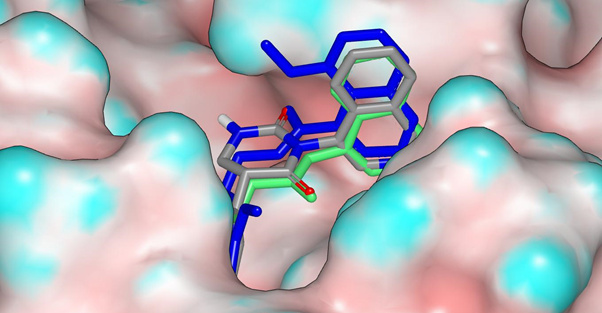
This view from a different angle shows more clearly how the carbonyl groups of the dihydrouracil overlay with the amide carbonyl groups of ADA-UCB-6c2cb422-1 (green) and EDJ-MED-6af13d92-3 (blue).