Topic automatically created for discussing the designs at:
https://covid.postera.ai/covid/submissions/PET-UNK-abc197b8
I’ll provide additional supporting information for the submission which is a variant of VLA-UCB-50c39ae8-3. The proposed binding mode was generated by docking the designed structure into the crystal structure of [ADA-UCB-6c2cb422-1] (x10959) (https://covid.postera.ai/covid/submissions/6c2cb422-4898-41bd-9409-adfdfb62f5c2/1) using hybrid followed by molecular mechanics energy minimization using szybki (both software tools are from OpenEye). I would expect the carbonyl oxygen atoms in the designed structure to be better hydrogen bond acceptors than the corresponding oxygen atoms in VLA-UCB-50c39ae8-3
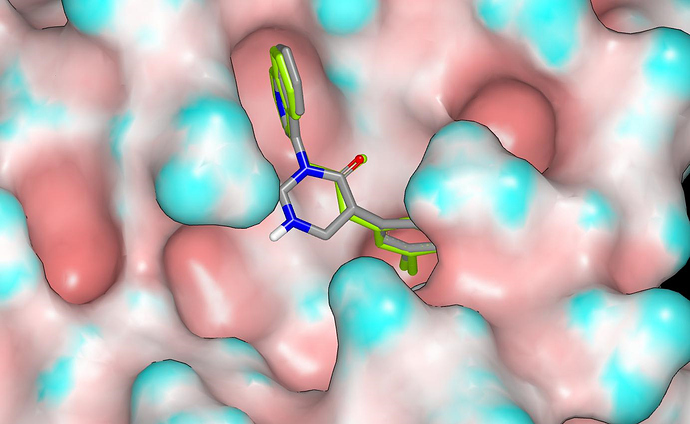
Here’s a graphic showing the proposed binding mode for the design with the crystallographic ligand in green (molecular surface of the protein has been colored by curvature)
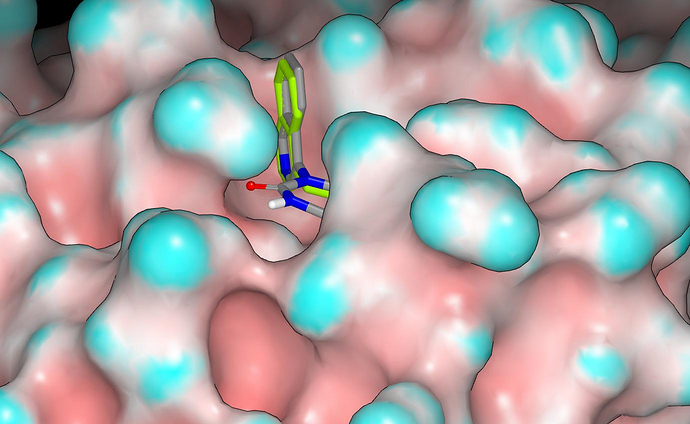
This is from another angle and it gives a better view of the proposed hydrogen bond between the carbonyl oxygen and G143.
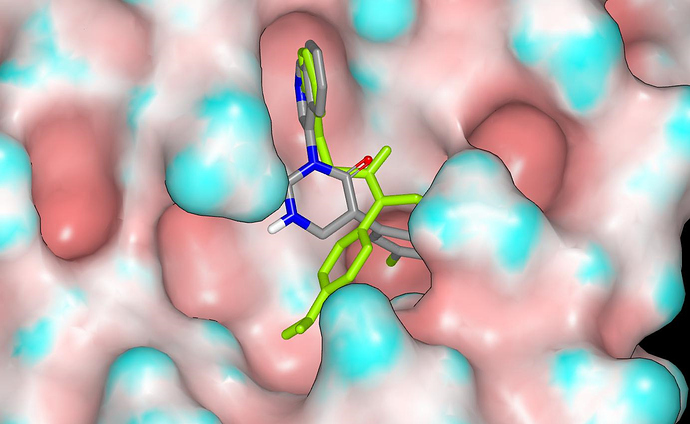
This shows the direction of the NH vector relative to the binding site and I’ve included ALP-POS-d2866bdf-1 (in green and taken from X10876 crystal structure). The NH vector appears to have potential for structural elaboration.
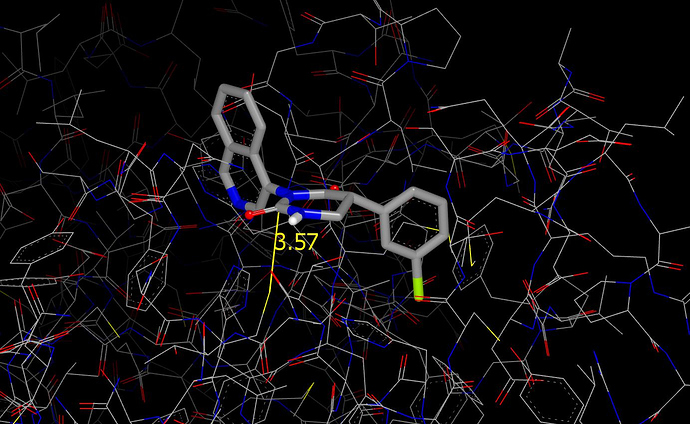
In the proposed binding mode, the carbonyl carbon is close to the thiol(ate) of the catalytic cysteine. This could result in what is sometimes referred to as an orthogonal dipolar interaction (this is not a term that I find particularly useful) or even turnover. I would expect a stronger interaction between thiol(ate) and carbonyl carbon for VLA-UCB-50c39ae8-3 than for this design.
This CSD crystal structure of a typical acyclic imide shows why an acetyl group on the amide nitrogen will adopt the wrong conformation for presenting the carbonyl oxygen to G143. Here’s a crystal structure from the CSD with the core substructure from the design.