Topic automatically created for discussing the designs at:
https://covid.postera.ai/covid/submissions/PET-UNK-94460c07
Designs in the PET-UNK-94460c07 submission are derived from the uracil PET-UNK-e8933450-1 which is intended to hybridize/merge the 3-aminopyridine-like series with the quinoline series while eliminating the dihydrouracil chiral center. This article presents indirect evidence that the uracil ring is recognized by the target. The hydrogen bond donor used by the quinolone amides (G143 backbone NH) is less solvent exposed than the hydrogen bond donor used by the 3-aminopyridine-like amides (E166 backbone NH). The designs in this submission substitute the uracil NH with methyl or ethyl and I’ve also included 3 variations for the P1 heteroaromatic substituent in order to provide options for the design team. I would anticipate that the potency advantage of isoquinoline over pyridine will be attenuated by the presence of two flanking carbonyl groups (they will force even pyridine out of coplanarity with the imide substructure) and am assuming that the P1 SAR would first be mapped using the parent uracil scaffold (unsubstituted NH).
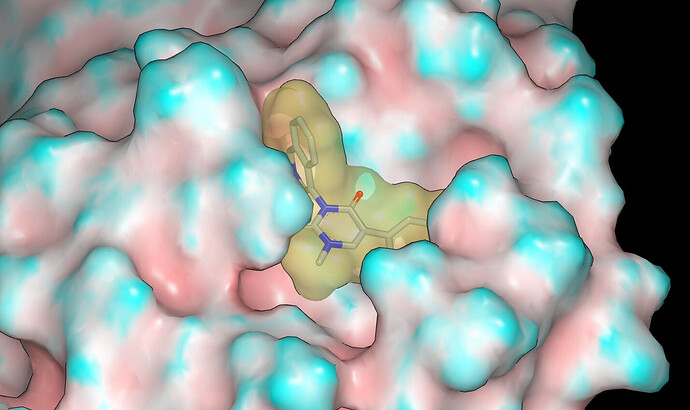
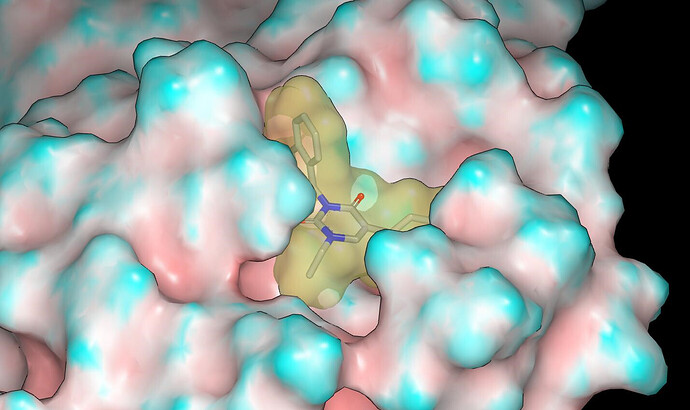
The binding mode was modelled using Szybki (OpenEye) and energy was minimized using the MMFF94S force field. The x10789 crystal structure (ligand: TRY-UNI-2eddb1ff-7) was used for modeling and I’ll mention @frankvondelft in case there is interest in how the crystal structures are being used in design. Based on consideration of contact with the molecular surface of the protein and the apparent lack of hydrogen bonds between the NHs of the parent compounds and the protein, I would anticipate potency in the order: NH < NMe < NEt.
Proposed binding modes for the designs are given in the pdb file associated with the submission. Here are graphics showing the binding mode modeled for PET-UNK-94460c07-1 (N-methyl) and PET-UNK-94460c07-2 (N-ethyl). The protein molecular surface is colored by curvature.