Topic automatically created for discussing the designs at:
https://covid.postera.ai/covid/submissions/fa25ac7f-b7ca-4ac6-8a32-5070858535ad
Due to a coding error on my part, the PDB submitted with this submission was not properly uploaded. I am providing it for download here!
DAV-AUT-fa2.pdb (472.2 KB)
I will try to fix this error with the site soon. Sorry!
Many thanks Davide. Very interesting designs! Thanks so much for putting the time into them! Just wanted to reply to your email here.
Please do share your synthetic routes. Happy to ask our chemists to look into that. Our platform suggests that the routes will be rather long (see attached for an example), thus can take weeks to make them. We have only sent molecules that are readily synthesizable from Enamine building blocks because we want to get assay results ASAP for this first round.
We will select more complex but promising molecules to synthesise when we close Wave 1 and Wave 2 tomorrow. Will definitely keep you posted!
DAV-AUT-fa2-4.pdf (114.4 KB)
Thank you for your input.
I will share the synthesis on the appropriate section of the “Make” section of the forum.
The molecules we designed for the challenge are fairly complex, requiring multiple steps synthesis. This was done on purpose. Fragment binding determination to the viral protease was done using mass spectrometry screening. A very sensitive technique for low affinity binders.
When we started the challenge, to our knowledge, there were no detailed information about the assay protocol for compound testing. We had to act quickly and decided to focus the design molecules which would have properties matching the profile of potential drug candidate (suitable for assays involving more complex biological systems). In mass spectrometry screening: potency, membrane penetration and metabolic stability are not relevant - in cell based assays, they might play a part in compound efficacy.
Please do not hesitate to contact us in order to get more information or discuss the design of the molecules.
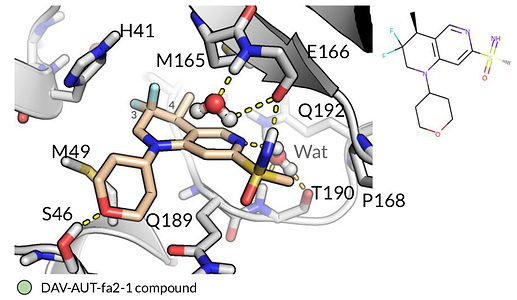
Sulfonamide of fragment X0161 can be substituted with a sulfoximine. A very versatile group that is able to interact with Glutamate 166 back bone. This nitrogen on the central tetrahydronaftiridine core can interact with the water molecule bridging Glutamine 192 and Threonine 190. The bigger central core, in particular the methyl group at 4 position, the gem-difluorinated methylene at the 3 position along with the tetrahydropyranyl moiety is able to occupy the binding pocket extending to Methionine 49 side chain. This group is engaging in an additional hydrogen bond interaction with Serine 46 side chain. This molecule is able to engage in several hydrogen bonds in the binding pocket, including bridging critical water-protein interactions.
This set of molecules, also ranked top 2% from the docking run that was reported here:
Synthesis for these compounds are reported in the “Make” section of the forum:
This work and project related activities reflect my own scientific vision, I am working pro bono, using vacation time and own resources (IP and computational equipment).