Topic automatically created for discussing the designs at:
https://covid.postera.ai/covid/submissions/PET-UNK-c5865d42
These structures came out of a discussion with @edgriffen and @JohnChodera on Ed’s submission EDJ-MED-50011917 and I’ll mention @mc-robinson and @alphalee in case they are interested.
I’ve generated potential binding modes for the azetidines PET-UNK-c5865d42-1 and PET-UNK-c5865d42-2. These came out of discussion of the EDJ-MED-50011917-1 design with @edgriffen @JohnChodera and I’ll include @mc-robinson. The azetidines were designed to donate a hydrogen bond to the backbone carbonyl oxygen of E166 and I’ve used the crystal structure (X10789) of TRY-UNI-2eddb1ff-7 (which has a beta-lactam substituent) for modelling. I’ll also mention @miko_a who is interested in targeting this region of the binding site (e.g. MIC-UNK-644c43c7). The related submission PET-UNK-b87f07d0 may also be of interest.
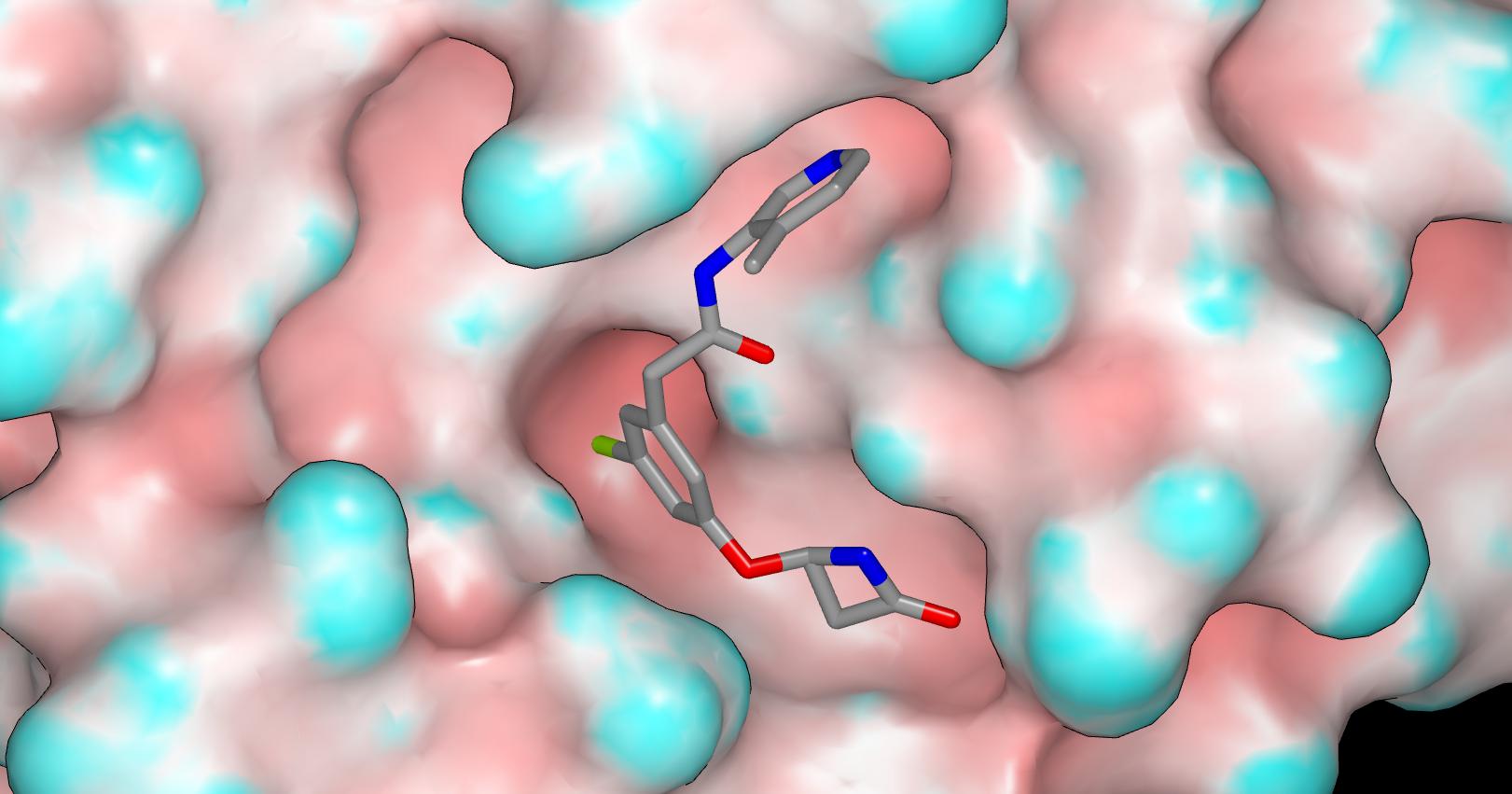
Here’s a graphic of TRY-UNI-2eddb1ff-7 in the X10789 crystal structure (molecular surface of the protein is colored by curvature).
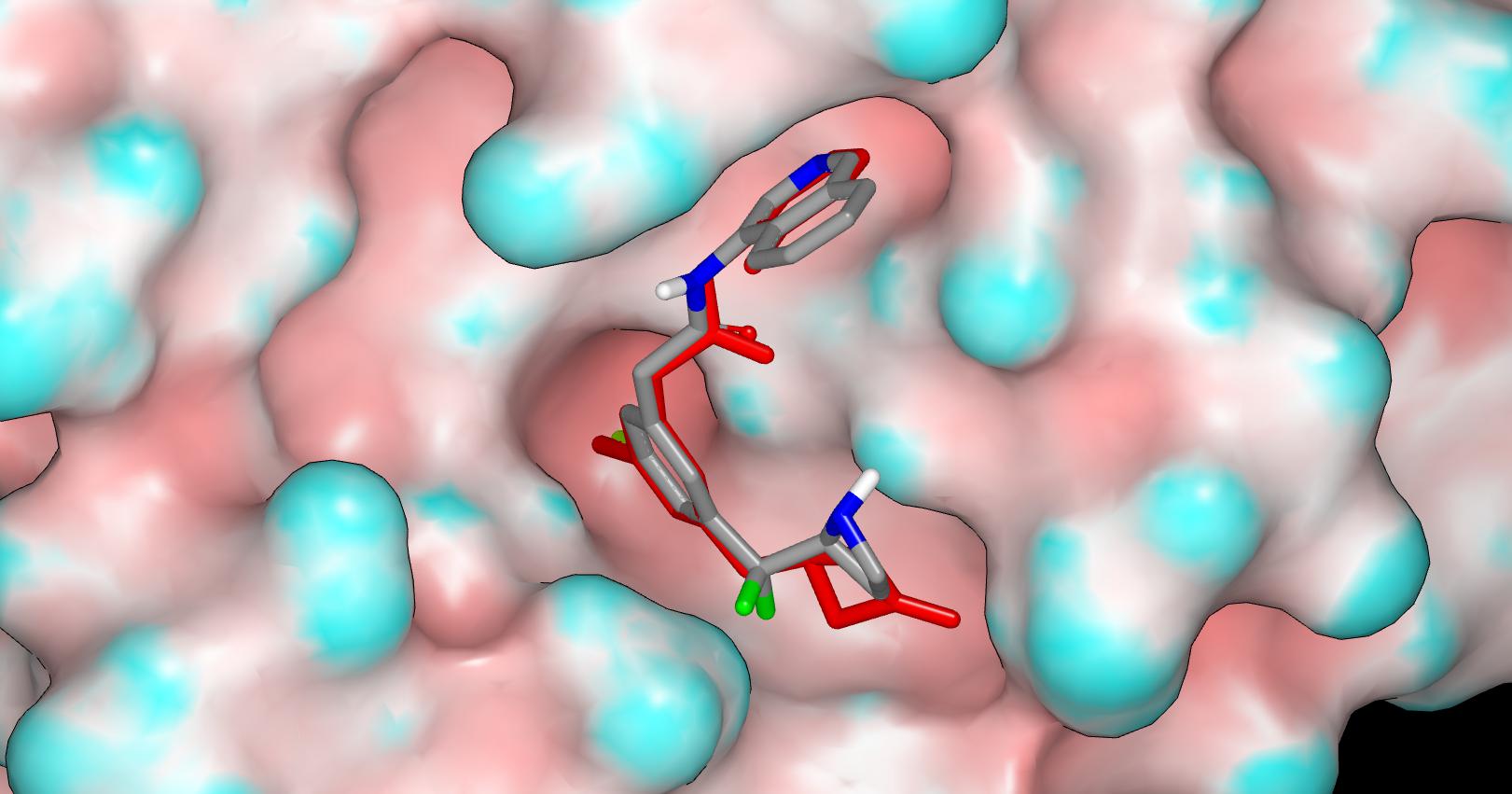
Here’s the proposed binding mode for PET-UNK-c5865d42-1 (the crystallographic ligand is shown in red) in which the design has been modeled in its protonated form.
Here’s the proposed binding mode for PET-UNK-c5865d42-2 (the crystallographic ligand is shown in red) and the design has been modeled in its neutral form since the inductive effect of the CF2 group is expected to push the azetidine pKa below physiological pH. As an aside, the electron-withdrawing nature of CF2 would also be expected to protect the ring from oxidation by CYPs to some extent.
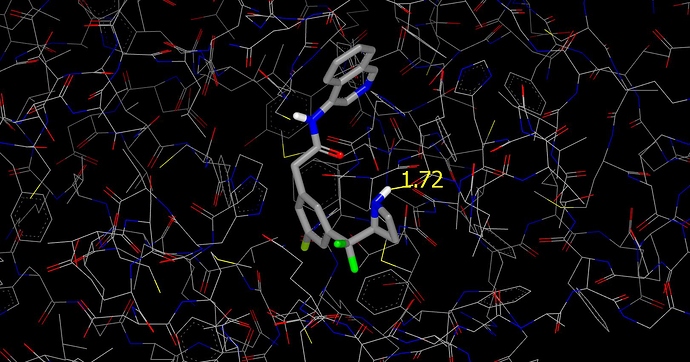
I’ve also generated a graphic without the surface so that you can see the hydrogen bond between the azetidine NH of PET-UNK-c5865d42-2 and the backbone carbonyl oxygen of E166.
The proposed binding modes can be accessed here azetidines_with_x10789.pdb (445.0 KB)