Topic automatically created for discussing the designs at:
https://covid.postera.ai/covid/submissions/PET-UNK-b87f07d0
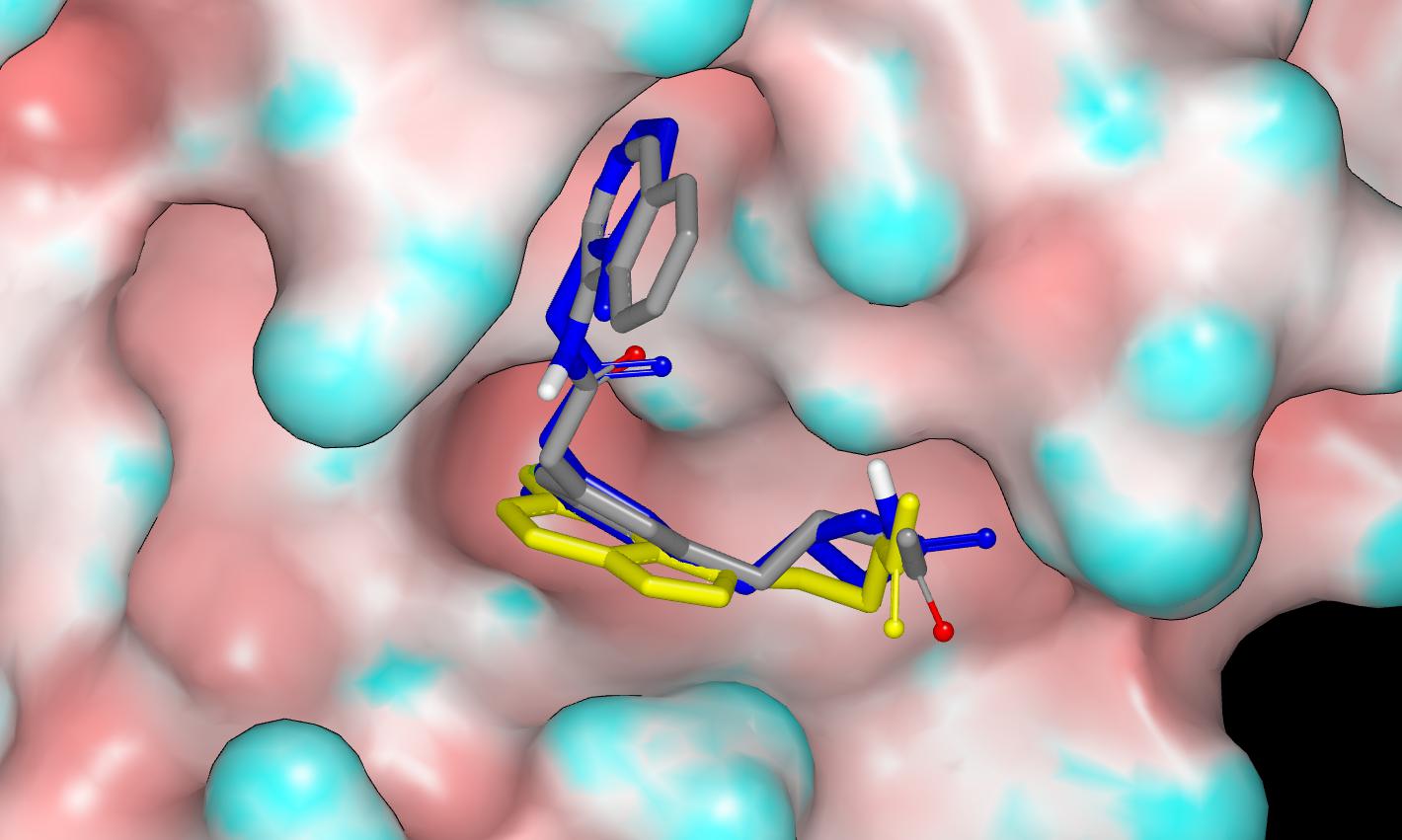
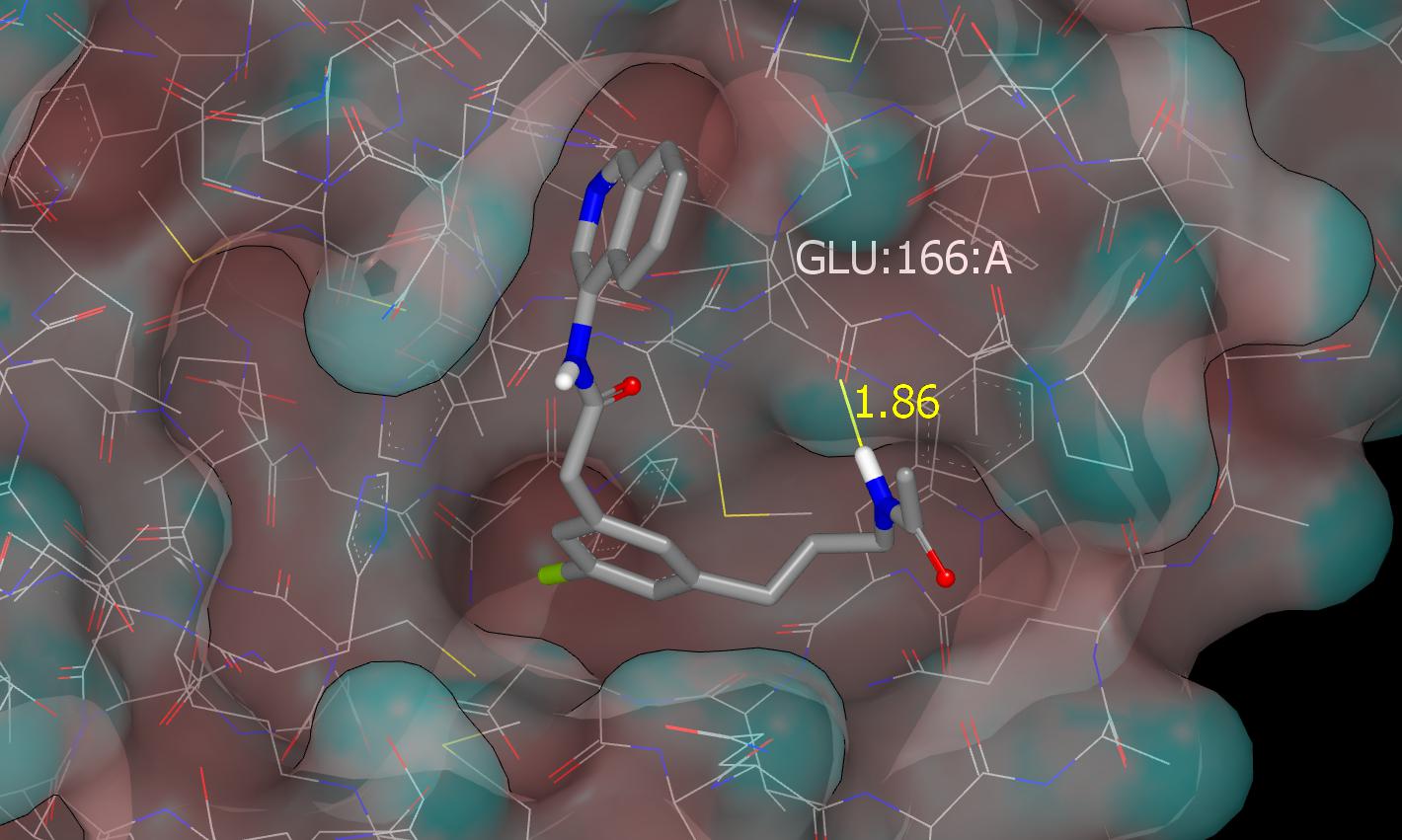
Like PET-UNK-c5865d42 this submission targets the backbone carbonyl oxygen of E166 and I’ll mention @mc-robinson @edgriffen @miko_a since they may be interested in exploiting this region of the binding site. The design tactic is to hybridize (merge) ADA-UCB-6c2cb422-1 with the fragment hit AAR-POS-d2a4d1df-2 and I’ll also mention @frankvondelft because these designs directly exploit the fragment screening output. The crystal structure (x10789) of TRY-UNI-2eddb1ff-7 was used for modeling and the binding mode of AAR-POS-d2a4d1df-2 observed in the x0104 crystal structure was used to guide the docking of PET-UNK-b87f07d0-1 which was performed by eye and refined by molecular mechanics (MMFF94) energy minimization.
The other three designs in the set have features that may prove beneficial. PET-UNK-b87f07d0-2 has a methyl group that should stabilize the bound conformation and may provide additional non-polar contacts with the molecular surface of the protein. The trifluoroacetyl group of PET-UNK-b87f07d0-3 should increase the hydrogen bond acidity of the amide NH and PET-UNK-b87f07d0-4 replaces the amide with a non-hydrolyzable analog.
Here’s a graphic (protein surface colored by curvature) showing the proposed binding mode for PET-UNK-b87f07d0-1 in the x10789 protein structure with the crystallographic ligand TRY-UNI-2eddb1ff-7 shown in blue and the fragment hit AAR-POS-d2a4d1df-2 shown in yellow.
This graphic shows the hydrogen bond between the acetamido NH and the backbone carbonyl oxygen of E166.
Is it not a bit dangly for a drug? Though yes, good spot, that x0104 does the same work as the beta-lactam of TRY-UNI-2eddb1ff-7.
(Here are direct fragalysis views:
Thanks for the fragalysis links, Frank, and a structure download was also provided with the original submission. I too see ‘dangliness’ as potentially undesirable and would generally try to form interactions as directly as possible. These comments on the PET-UNK-c5865d42 submission might be of interest.
I see the beta-lactam as also being potentially undesirable given that it is what might be called a ‘gratuitous electrophile’. Comparison of IC50 values for ADA-UCB-6c2cb422-1 and ALP-POS-3b848b35-2 indicate that potency gains (ranging from 2.7 in the fluorescence assay to 6.6 in the RapidFire assay) resulting from inclusion of the beta-lactam substructure are modest. The IC50 ratio for the fluorescence assay is actually less than the ratio of 3.6 that @Wal-Ward has recommended for being at least 95% confident that an observed potency difference is real.
The betalactam is a metabolic disaster and we’re desperately trying to get rid of it.
Given that my involvement with the project is peripheral, it would not be prudent for me to use terms like ‘disaster’ to describe project compounds. I’ll just suggest that the beta lactam represents a Pyrrhic victory in the optimization campaign. I’m working on a couple of other options for targeting E166 and will submit designs if any of these appear to have potential.
It also is more Pyrrhic in terms of protease selectivity, or more a an “opportunity in poly-protease space” as I’m sure the more entrepreneurial would describe it…
A potency change up to and including the minimum significant ratio or MSR is likely to be due to noise (estimated probability 95%). For the fluorescence assay, I calculated the MSR as 3.6 (see brief report). I now can add that my estimated MSR for Rapid Fire is 3.1 (based on 13 data sets for CVD1643). I suggest that MSRs are considered when analysing whether an observed activity change is likely to be real or only in the noise.
Thanks for confirming this, Wal, and for providing the estimated MSR for the RapidFire assay.
Hi Ed, I really should have used ‘Pyrrhric victory’ in relation to this graphic from the Nature of Ligand Efficiency.