Nice, should be a good starting point
Nir, should we try again to get them into structures? Diamond is back up today, and we’ll be doing more Mpro experiments.
Dear All,
I would first like to commend this entire effort. It really is remarkable what has already been accomplished and the plans ahead.
In regards to this “Second design call” and from reading through this full thread, here are my thoughts:
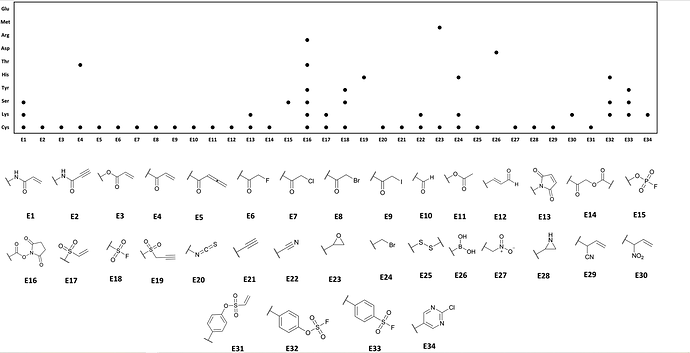
Considering the initial hits involved chloroacetamides, geometrically it would make the most sense to switch to fluoracetamides (weakly reactive, smaller than the Cl) however largely different reactivity so potency may take a hit. There are concerns of completely changing the warhead, however there are several options (see image below from a review article we just published).
If the acrylamides are not hits (possibly due to a lack of sufficient reactivity), we can move to a more reactive variant (Vinyl sulfonamides). Or the propiolamide seen in Acalabrutinib.
A second strategy (if we have the space within the pocket), we can utilize a technique typically seen in covalent kinase inhibitors. Whereby you use a poorly reactive warhead (i.e. acrylamide) and append a basic nitrogen group (dimethylamine) to this electrophile. This basic group accelerates Cys reactivity by possibly creating the thiolate nucleophile. It also acts as a good solubility handle (improving PK). See examples such as Afatinib.
Another approach, I have seen the use of 2-fluoropyridines are good warheads which are Cys reactive via a Nucleophilic aromatic substitution mechanism.
So that would be my suggestion, either:
- Fluoroacetamides
- More reactive variants of acrylamides (acrylamides with dimethylamines appendages)
- More reactive variants of acrylamides (vinyl sulfonamides)
- More reactive variants of acrylamides (propriolamides)
- SnAr-reactive variants (2-fluoropyridines)
I do have covalent docking capabilities as well and would be more than happy to run some calculations if that would be helpful with these proposed structures. I am just having trouble finding the two fragments of interest.
Lit: https://pubs.rsc.org/en/content/articlehtml/2020/cs/c9cs00720b
So my magic resident geek fxed it today - it really is better this way, thanks for the kick in the backside. Go Rachael Skyner!
@pgz I have now updated the list, any of the submissions that mentions a covalent fragment will now have a covalent flag. Note that this is prone to false positives if people submit many molecules (some of which are not based on covalents) as a part of a bigger submission.
Hi Frank,
I went to check the updated version and now it seems buggy - I’m getting to the fragalysis site via the links here:
https://www.diamond.ac.uk/covid-19/for-scientists/Main-protease-structure-and-XChem/Interactive-views.html
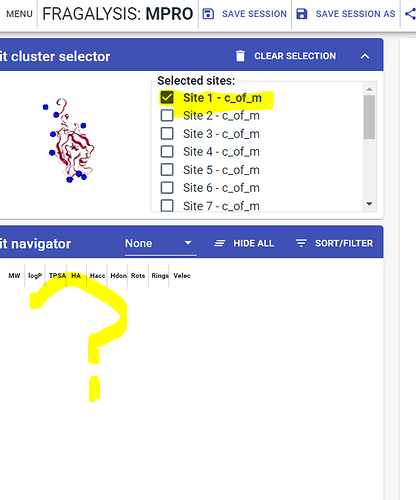
When it opens up all of the compounds from sites 1, 11 and 8 docked, and the list in the hit navigator. If I then unselect these sites in the cluster selector, and then reselect a site from the hit cluster selector, nothing shows up in the hit navigator (see screenshot below). Also I can’t see how the different fragments have been classified as covalent or not… all I see if that the sites have been renamed with an additonal “C_of_m” after each site
Is it possible that I’m not following the correct link to the fragalysis snapshot?
Hi all
This is a most welcome escape from online teaching and recording lectures and workshops and re-organising exam papers! Thanks!
Wow, a nice set of compounds for testing. Well done on that. Red on the Excel chart, I guess (many of mine included!) means not good? For what reason? Poor docking? Mine were by eye as I think our Glide expired and I don’t have a licence at home and I’m pretty hopeless with docking anyway!
Just a couple of additional points, perhaps if you have some really nice Cys-covalent dockers (caveat, you’ve probably already got 1 million +1 things already to contend with!);
SO2F and similar warheads e.g. Synfacts 2019; 15(12): 1436; *ACS Med. Chem. Lett. 2018, 9, 7, 584-586
oxoboroles (Benkovic/Anacor type) might be useful warheads to envisage:
DMARDs are interesting compounds and might warrant an investigation e.g. Leflunomide, especially its active metabolite: https://en.wikipedia.org/wiki/Teriflunomide?? Probable Michael acceptor, small and repurposable?
Thinking beyond in vitro, in vivo, a number of thiophenes and the like can be bioactivated to form a S-S link (via thiophene oxidation, ring opening). Think of Plavix and omeprazole? I know these won’t show up in a frag screen but, if you get hits, incorporating bioactivated warheads might be useful?
Comments now on the fragments are included.
I’d just be careful with some of the ureas (solubility) and a number of CF3 are very high logP. If these were fragments they’d be very poor leads in terms of LipE.
Anyway, comments attached. Please keep me posted. Well done-you all deserve a medal or some kind of recognition in acting on trying to make the world a safer place!
Stay safe.
Thanks
Best Wishes
John
Ben - this is now fixed - some postdocs did quick work this afternoon. Thanks for pointing it out.
Just joined and I hope that I’m posting in the right place. Here are some notes on the relevance of reversibility to the design of inhibitors of cysteine proteases:
My view is that reversible covalent inhibition is likely to be the easier option unless there is a compelling rationale for irreversible inhibition.
“A comparative study of warheads for design of cysteine protease inhibitors” may be useful if you’re thinking about reversible covalent inhibitors. Nitriles and aldehydes are well known warheads and, in the article, we introduced the oxime (potency intermediate between aldehyde and nitrile while providing access to the prime side of the catalytic site). There also some data for some azadipeptide nitriles which are very effective warheads (I wouldn’t claim to fully understand why). Here is the link: https://doi.org/10.1016/j.bmcl.2017.10.002
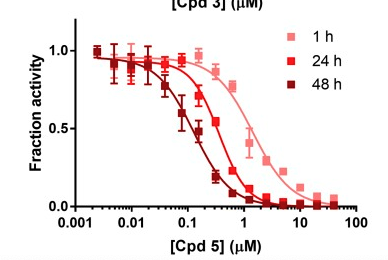 This is an example of an acrylamide covalent inhibitor that is an NNRTI, it takes up to 48h incubation to produce the max effect, assays are at different time periods, but also the molecule is probably a decent lead at 0.14uM while being probably about the size of two coupling reactions we could do with Enamine. Just to make the point that for these compounds, in the assay, we need to incubate for a series of time points to see if the trend fits covalent activity. Probably this has been thought about but thought I’d just point it out anyway. The other thing maybe to think about is to incubate with glutathione, we don’t want the compound being completely scavenged or causing liver damage. Proc Natl Acad Sci U S A. 2017 Sep 5; 114(36): 9725–9730.
This is an example of an acrylamide covalent inhibitor that is an NNRTI, it takes up to 48h incubation to produce the max effect, assays are at different time periods, but also the molecule is probably a decent lead at 0.14uM while being probably about the size of two coupling reactions we could do with Enamine. Just to make the point that for these compounds, in the assay, we need to incubate for a series of time points to see if the trend fits covalent activity. Probably this has been thought about but thought I’d just point it out anyway. The other thing maybe to think about is to incubate with glutathione, we don’t want the compound being completely scavenged or causing liver damage. Proc Natl Acad Sci U S A. 2017 Sep 5; 114(36): 9725–9730.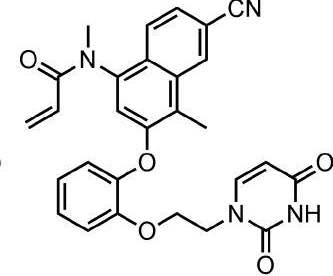
Just to point out that alpha-keto-amides will be also welcome to “qualify” for the fast track as successful examples that made it to FDA approval are available:
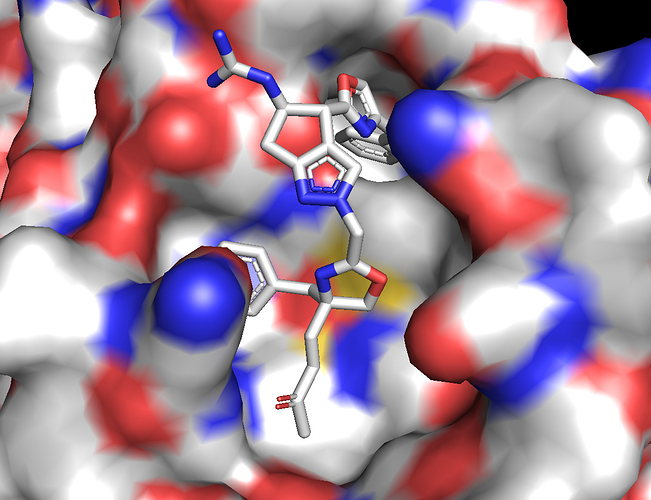
We have submitted a peptidomimetic molecule capable of forming covalent bond through a Michael acceptor [ABH-UNI-fbf]. The peptidomimetics contains two Phe residues which sit very nicely in two different hydrophobic pockets and an Arg which forms a salt-bridge to E166. The ligand buries a total surface area of 500 Å^2. The Michael acceptor is placed close to the Cys145 for covalent bond formation.
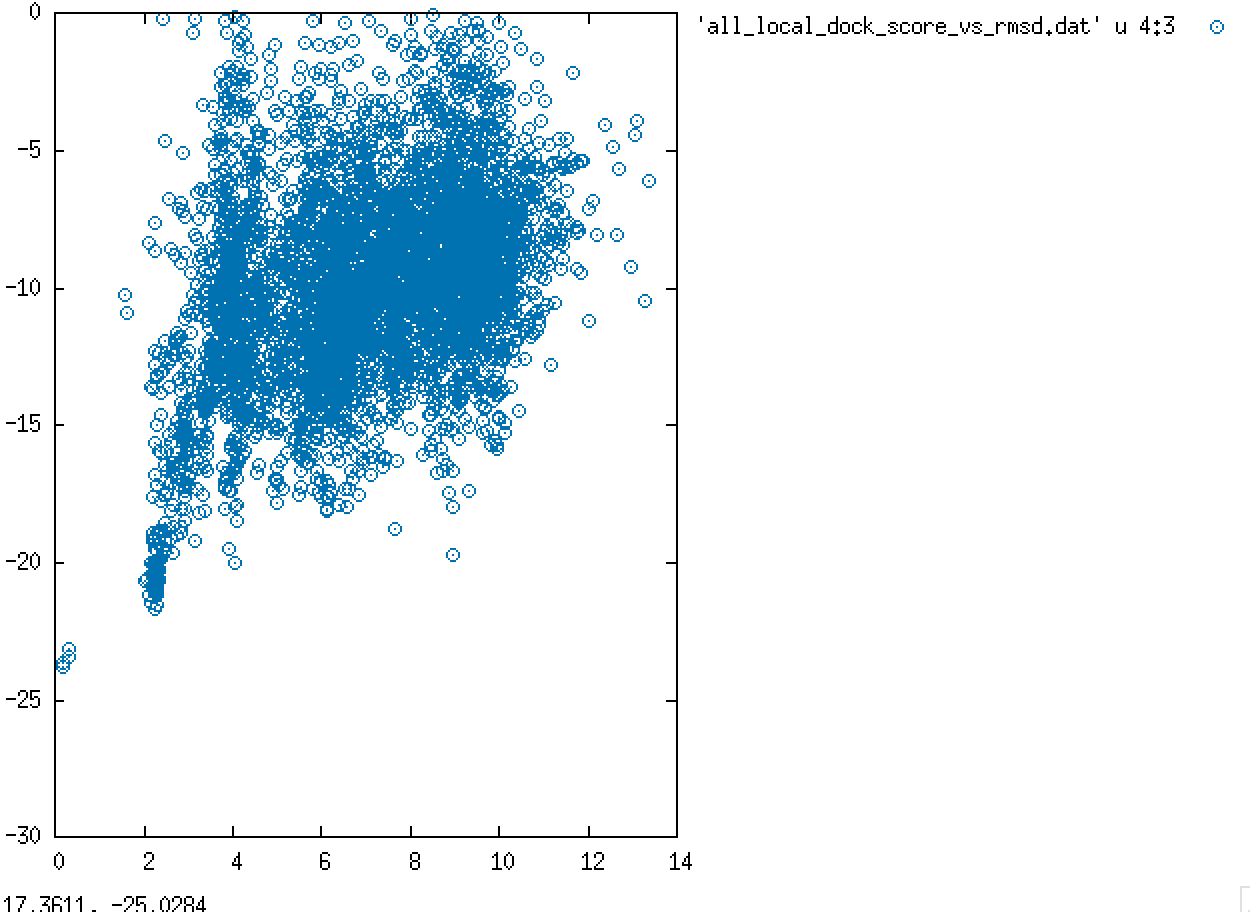
.We performed ligand docking in Rosetta using first a global search with 10k poses and then a refined search using the best pose from global docking as starting point. A plot of ligand RMSD vs interface_delta score (which is a measure of the surface area buried upon ligand binding) is shown below.
best wishes,
Abhishek
Thanks!
Which submission is it?
I thought I already submitted it but there was no conformation page afterwards. In any case here is SMILES
CC(=O)C=CC1(Cc2ccccc2)COC(Cn2cc3c(n2)CC(NC(N)=[NH2+])C3c2nc(Cc3ccccc3)co2)=N1
Maybe try submitting it again, make sure you add all the information needed.
Trying now. Some problem with the server, I guess.