Ben - this is now fixed - some postdocs did quick work this afternoon. Thanks for pointing it out.
Just joined and I hope that I’m posting in the right place. Here are some notes on the relevance of reversibility to the design of inhibitors of cysteine proteases:
My view is that reversible covalent inhibition is likely to be the easier option unless there is a compelling rationale for irreversible inhibition.
“A comparative study of warheads for design of cysteine protease inhibitors” may be useful if you’re thinking about reversible covalent inhibitors. Nitriles and aldehydes are well known warheads and, in the article, we introduced the oxime (potency intermediate between aldehyde and nitrile while providing access to the prime side of the catalytic site). There also some data for some azadipeptide nitriles which are very effective warheads (I wouldn’t claim to fully understand why). Here is the link: https://doi.org/10.1016/j.bmcl.2017.10.002
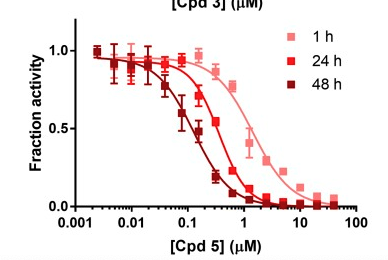 This is an example of an acrylamide covalent inhibitor that is an NNRTI, it takes up to 48h incubation to produce the max effect, assays are at different time periods, but also the molecule is probably a decent lead at 0.14uM while being probably about the size of two coupling reactions we could do with Enamine. Just to make the point that for these compounds, in the assay, we need to incubate for a series of time points to see if the trend fits covalent activity. Probably this has been thought about but thought I’d just point it out anyway. The other thing maybe to think about is to incubate with glutathione, we don’t want the compound being completely scavenged or causing liver damage. Proc Natl Acad Sci U S A. 2017 Sep 5; 114(36): 9725–9730.
This is an example of an acrylamide covalent inhibitor that is an NNRTI, it takes up to 48h incubation to produce the max effect, assays are at different time periods, but also the molecule is probably a decent lead at 0.14uM while being probably about the size of two coupling reactions we could do with Enamine. Just to make the point that for these compounds, in the assay, we need to incubate for a series of time points to see if the trend fits covalent activity. Probably this has been thought about but thought I’d just point it out anyway. The other thing maybe to think about is to incubate with glutathione, we don’t want the compound being completely scavenged or causing liver damage. Proc Natl Acad Sci U S A. 2017 Sep 5; 114(36): 9725–9730.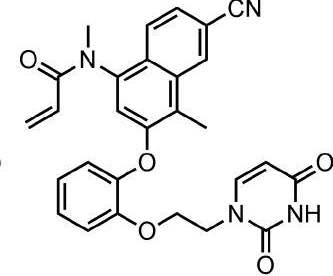
Just to point out that alpha-keto-amides will be also welcome to “qualify” for the fast track as successful examples that made it to FDA approval are available:
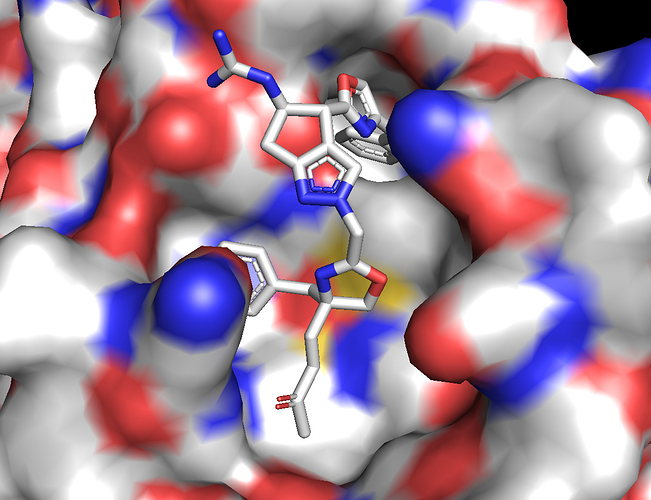
We have submitted a peptidomimetic molecule capable of forming covalent bond through a Michael acceptor [ABH-UNI-fbf]. The peptidomimetics contains two Phe residues which sit very nicely in two different hydrophobic pockets and an Arg which forms a salt-bridge to E166. The ligand buries a total surface area of 500 Å^2. The Michael acceptor is placed close to the Cys145 for covalent bond formation.
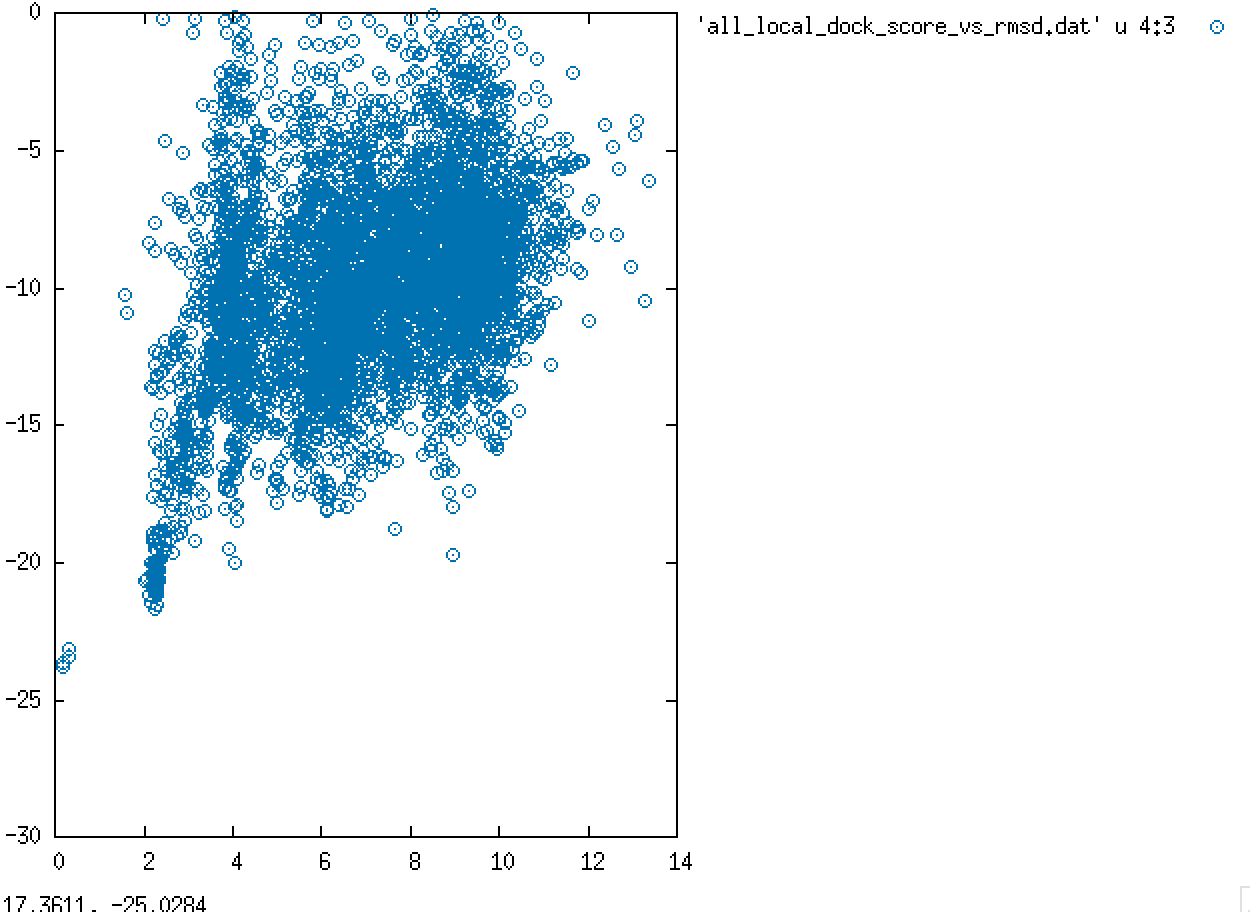
.We performed ligand docking in Rosetta using first a global search with 10k poses and then a refined search using the best pose from global docking as starting point. A plot of ligand RMSD vs interface_delta score (which is a measure of the surface area buried upon ligand binding) is shown below.
best wishes,
Abhishek
Thanks!
Which submission is it?
I thought I already submitted it but there was no conformation page afterwards. In any case here is SMILES
CC(=O)C=CC1(Cc2ccccc2)COC(Cn2cc3c(n2)CC(NC(N)=[NH2+])C3c2nc(Cc3ccccc3)co2)=N1
Maybe try submitting it again, make sure you add all the information needed.
Trying now. Some problem with the server, I guess.