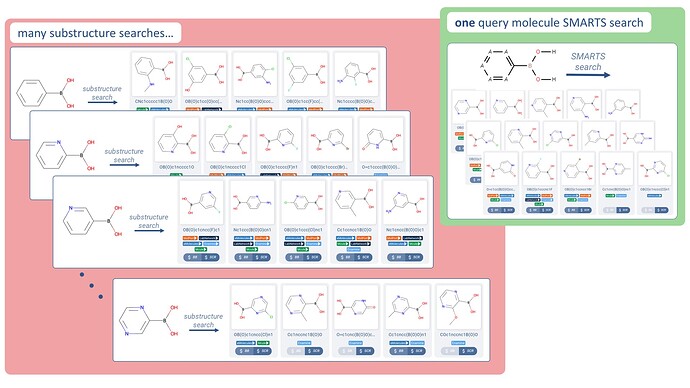
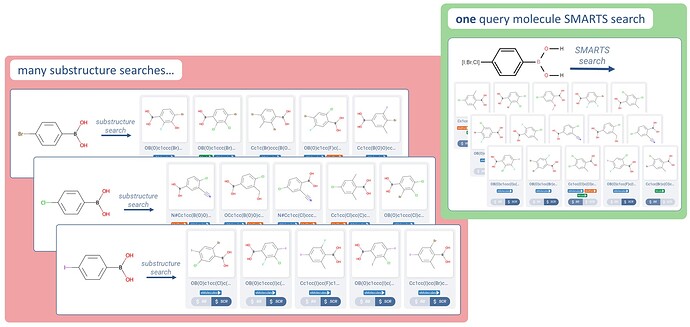
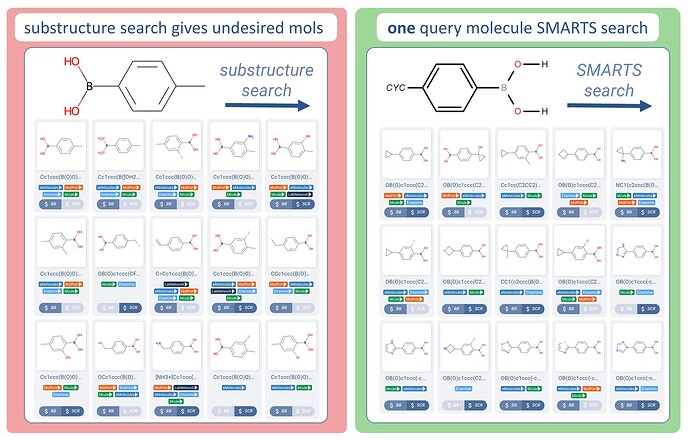
TL;DR - Query molecules are powerful and now in Manifold.
Query Atoms - Search for structures with a predefined set of atoms at a desired position
Example: encode the many substructure searches for substituted benzene and pyridyl rings
→ Gives you all heteroatom permutations of this aryl boronic acid.
Query Atom Lists and Not-Lists - Search for structures with (or without) a custom set of atoms at a desired position
Example: get all molecules with a chlorine, bromine, or iodine at a given position
→ Returns all chloro-, bromo-, and iodo- functionalized boronic acids.
Query Groups - Use predefined generic groups to include in hits
Example: show me a variety of intermediates already coupled to a second ring
→ Returns a large array of interesting boronic acids already coupled to a cyclic group.
Searching with Manifold
When designing new synthetically accessible candidates, a chemist might first look to their inventory or the inventory of building block suppliers to explore the chemical space around one of the building blocks used in a robust synthesis strategy. Ideally, they would like to hold parts of the original building block constant, while varying other groups strategically.
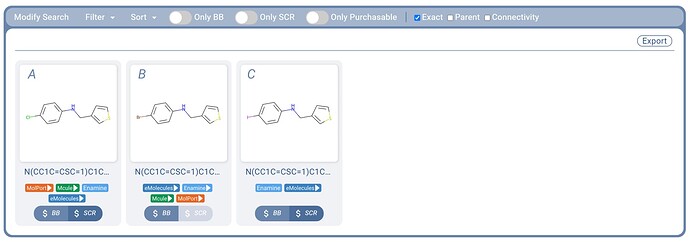
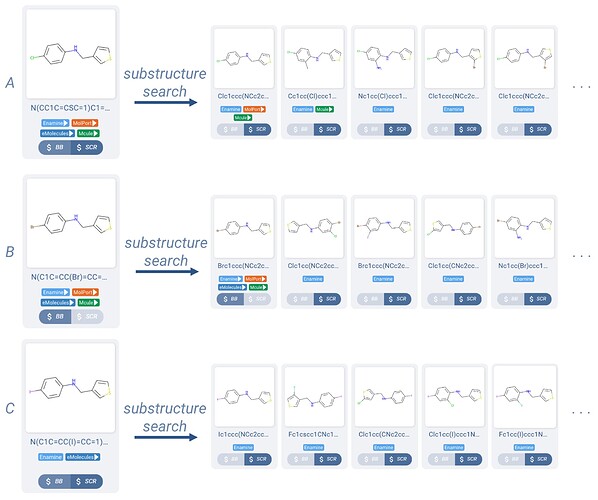
A common example of this would be taking the amine building block shown Figure 1.A, allowing the chlorine to be either chlorine, bromine, or iodine, and finding building blocks which contain that building block as a substructure.
Figure 1. A Manifold search query with three halide variations of the same building block.
In Manifold, the chemist could load a search query (Figure 1) with the three variations of the original building block, and then explore substructure matches for each in sequence. Some of the results returned for each are illustrated in Figure 2.
Figure 2. A sampling of substructure results for each of the three molecules in the search query of Figure 1.
These results are a great start, as there are many hits which could be interesting substitutions to the original amine. However this mode of searching will require that all hits contain the exact substructure, not allowing for substitution of the atoms on the ring, or varied groups at all. The branching off of the substructure is also not controlled, allowing for multiple halide or even primary amine substituents off of the benzene, the latter of which will lead to selectivity issues upon using this building block in amide-coupling chemistry.
As illustrated, there are two glaring limitations to this approach:
- Doing multiple searches on slightly varying building blocks is repetitive and not as efficient as it could be.
- A substructure search leaves much to be desired when it comes to building blocks with more interesting variations from the original; we are stuck with a benzene and a thiophene ring in every molecule
Thankfully, SMARTS searching allows us to tailor a single query to get closer to the exact chemically diverse space desired.
Cue Query Molecules 
Unless you are fluent in the SMARTS language, you are probably unaware of a pretty powerful search mode in Manifold – SMARTS searching. An extremely feature-rich, programmatic method for querying molecules, the SMARTS language by Daylight Information Systems is a household name for a cheminformatician.
Albeit powerful, the SMARTS language is tricky to work with. Through this frustration, many systems for working with SMARTS graphically were born, to enable crafting so-called “query molecules”. Two of the most well-known graphical SMARTS tools are: (1) ChemAxon’s powerful JChem office extensions, and (2) SMARTS.plus from the Universität Hamburg.
We realize that Manifold would not be complete without its own query molecule interface, and as such, is now equipped with an effective graphical SMARTS query feature. This feature builds upon the open-source web-based molecule drawing tool, Ketcher, and enables our users to design query molecules using the following components:
- Atom lists and atom not lists
- Query atoms
- Generic query groups
Let’s take the example amine building block from above to illustrate these query molecule features.
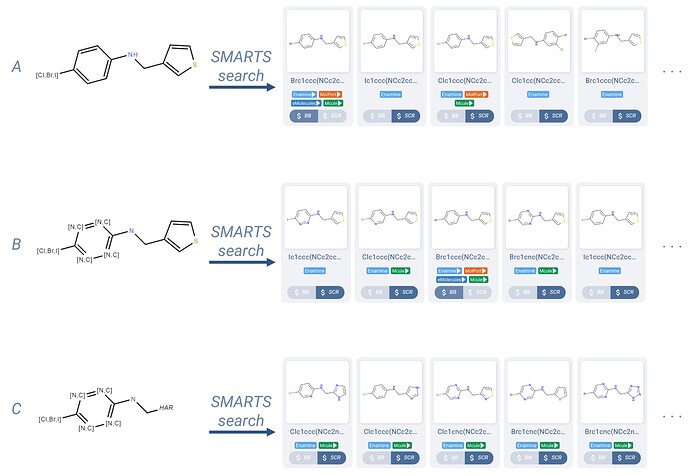
Figure 3. Three query molecule examples and a sampling of their SMARTS search results. All examples make use of the atom lists feature, and example C also uses a generic HAR group (heteroaromatic).
An atom list of [Cl,Br,I] used inplace of a single halogen atom (Figure 3.A) allows us to obtain the same hits that took three substructure searches previously (Figure 2), with a single query molecule SMARTS search.
Another use of atom lists can be seen in Figure 3.B, where we allow the benzene ring to be heteroatomic by allowing the four carbons not participating in a substituent bond to be either nitrogen or carbon ([N,C] atom list).
We can further curate the set of results by allowing the thiophene to be any heteroatomic aryl group. This is done with the use of a generic query group, in this case HAR (heteroaryl). As shown in Figure 3.C, the results are quite diverse, while maintaining required structural and atom features.
Of course these queries could be performed directly in Manifold using SMARTS, but this tooling allows a SMARTS-novice to access much of its power, and avoid writing things like:
[#6]1(:[#6,#7]:[#6,#7]:[#6](:[#6,#7]:[#6,#7]:1)-[#35,#17,#53])-[#7]-[#6]-;!@[a;!$(c1ccccc1);!$(c1cccc1)]
Library Chemistry with Query Molecules
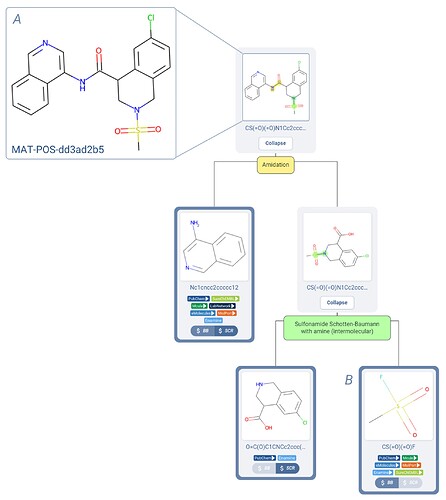
Now that I’ve got you hooked on query molecules, let’s take things a step further and deploy this search mode within a real campaign! For example, let’s consider this compound as was proposed to the COVID Moonshot campaign in submission MAT-POS-dd3ad2b5 as an example starting point, and the Manifold-proposed 2-step synthetic route utilized in this series (Figure 4).
Figure 4. A COVID Moonshot submission (A) with a robust and synthetically accessible retrosynthesis route from Manifold (B).
If possible, you would really like to stick with this synthetic strategy, as your CRO favors this method, and has plenty of the acid and amine building blocks in-stock. However, based on recent data, the med chem team would like to vary the sulfonyl halide to improve the metabolic stability of the lead while gaining or maintaining potency.
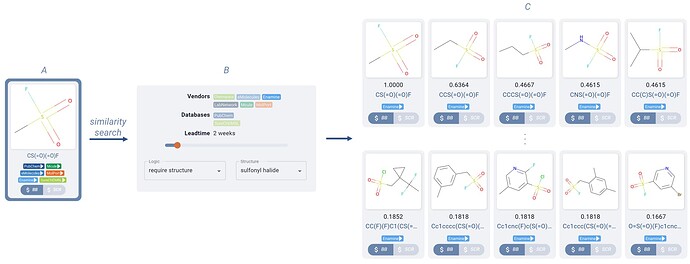
Clicking-through the sulfonyl fluoride building block (Figure 5.A) on the route, you search through the similarity hits that Manifold suggests (Figure 5.C), filtering down to hits which are available off-the-shelf only from Enamine, and applying a boolean filter (to learn more about boolean filtering in Manifold, checkout this blog post) to require the presence of a sulfonyl halide group (Figure 5.B).
Figure 5. Sulfonyl chloride similarity search hits (C) for the sulfonyl halide building block (A) of Figure 4B. Results are filtered down to two week lead time, only in Enamine catalogs, and required to contain a sulfonyl halide required structure via boolean filters (B).
Of the many results returned, there are indeed some interesting analogs,which could be used to build out a diverse library. However, to produce a library with more control over the chemical environment of this moiety, the following constraints would like to be satisfied:
- Maintain at least one methylene carbon between the sulfone and the new additions to the building block
- Require the presence of either an ether or a nitrile electron-withdrawing group
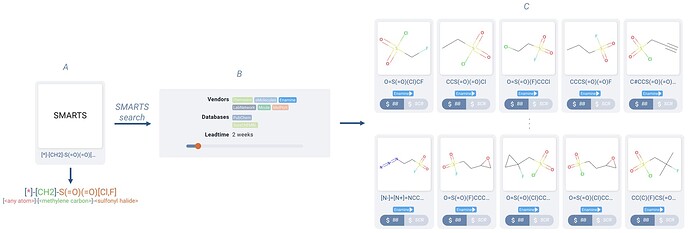
An example SMARTS query to explore the generic sulfonyl halide space would be [*]-[CH2]-S(=O)(=O)[Cl,F], which would match the general structure: [<any atom>]-[<methylene carbon>]-[<sulfonyl halide>] (Figure 6.A).
Figure 6. Example SMARTS search hits (C) for a query molecule which encodes a desired set of features in the resulting matches. This query (A) encodes the requirement for the presence of a sulfonyl halide group, as did the similarity search in Figure 5, but now without the need for boolean filters. Results are filtered down to two week lead time, only in Enamine catalogs (B).
As a SMARTS master, you would be able to expand out this query to capture the exact building block sets desired. But for those of us who would prefer to craft queries graphically, the new query molecule features in Manifold can help us here yet again.
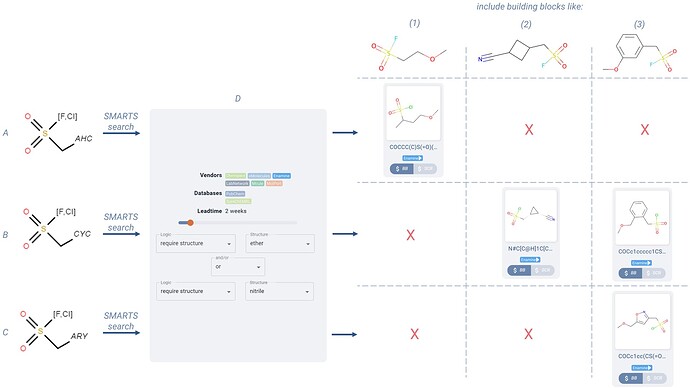
To focus only on sulfonyl fluoride and sulfonyl chloro building blocks, we can use an atom list to require [F,Cl]. To keep our methylene carbon, we can just use a single carbon bond to add this to the query. All three example query molecules in Figure 7 (A-C) maintain these two query features. We can then use boolean filters to require the presence of an ether OR a nitrile functional group within the hit molecules (Figure 7D).
With these baseline constraints in mind, you would also like to systematically investigate three different sets of building blocks, differentiated as follows, and illustrated in the examples on the top right of the table in Figure 7 (1-3)
- Acyclic moieties
- Building blocks with an aliphatic ring
- Building blocks with an aromatic ring
To get such specified groupings of building blocks, it is important to constrain for desired functionality, while making sure to exclude others. We can use query molecule generic groups to narrow in on these sets.
Query of Figure 7.A generally captures building blocks without rings (using the acyclic general group AHC), capturing capturing less rigid building blocks.
The Figure 7.B query very broadly captures ring-containing analogs (with CYC generic group) like Figure 7 (2-3), whereas tweaking the query slightly, allowing only aromatic cyclic moieties (with ARY generic group) off of the methylene carbon, narrows in your results on things like Figure 7.3 only.
Figure 7. For the three query molecule rows (A, B, C), if the SMARTS results would return building blocks like those displayed on the top of each column (1, 2, 3), then an example hit is displayed. Results are filtered down to two week lead time, only in Enamine catalogs, and boolean filters are used to require either an ether or a nitrile structure (D).
You will notice that none of the three query molecules can successfully yield building blocks of the 2nd category, without also including those in category 3, as a CYC generic group will capture both aromatic and aliphatic rings.
Using a combination of query molecules will allow you to tailor a set of building blocks to build out a library to make improvements to the metabolic stability of the hit, but the specific categories desired need to be acquired via the SMARTS language directly.
Why you should still learn SMARTS
While the results using Manifold graphically will get you nearly there, the control you have by using the SMARTS language directly is unmatched. Two such limitations are: (1) generic groups cannot be chained together, and (2) you cannot embed logical statements for required/excluded generic groups. One example SMARTS which is not possible graphically would be:
[<sulfonyl halide>]-[<HAR or CYC> && <not 6-membered ring>]-[<nitrile>]
Beyond the scope of this post are even more intricacies of the SMARTS language, including
- Recursive SMARTS - an expression which allows you to define atomic environments, and can even be nested for more programmatic control of results.
- SMARTS logic - powerful boolean logic to include and exclude groups at the precise position of interest.
- Reaction SMARTS - a nomenclature which enables a chemist to define the transformation of atoms in a chemical reaction.
Query molecule searching is currently offered for free on the Manifold platform, so we hope you give it a try!