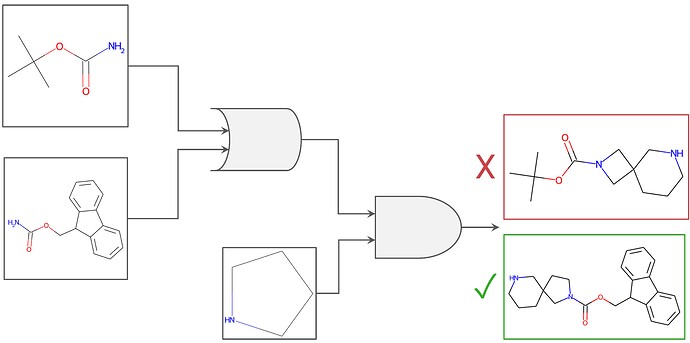
Figure 1. Graphical representation of boolean logic as applied to chemical building block filtering. Filters to require an NBoc OR an NFmoc protecting group, AND required the presence of the custom-drawn cyclic amine were used. Applied to a similarity search, for example, these constraints would filter out the result on the top right (the cyclic amine desired is not present, although the NBoc group is), whereas the constraints would include the bottom right result (presence of NFmoc AND the desired cyclic amine).
Have you wanted ‘similar’ to mean something slightly different while performing a similarity search? How about wanting your substructure search to only return matches with specific protecting groups? Or maybe exclude from your search results structures which trigger medicinal chemistry alerts?
I bet you’ve nodded your head at one of the above. And I would also bet that you had to go to your cheminformatics expert (or code up a script yourself) to find solutions to these common search problems. Or perhaps you have found some enumerated libraries which suit your needs - although these often have downsides of being rigid, outdated, etc.
To solve these search problems, Manifold now allows users to integrate logical queries into similarity and substructure searching. Our goal is to enable any chemist to “code” their own detailed queries, without feeling the need to find a programmer to help them.
This is facilitated by integrating the ideas of boolean logic directly into the search tool. Conditional “IF… THEN logic” is at the heart of most computer programs, as well as many medicinal chemistry tasks. For example, a med-chemist might often be use logic like:
IF molecule does not contain PICK AN UGLY FUNCTIONAL GROUP
THEN include in screening library
Or maybe even some more complex logic for assembling a parallel chemistry building block library:
IF molecule has a CARBOXYLIC ACID
AND molecule has an NBoc PROTECTED AMINE
AND molecule DOES NOT have an UNPROTECTED PRIMARY AMINE
THEN add to parallel library building blocks
Or a med-chemist might use some logic like the following to determine if their molecule is lead-like:
IF molecule has MW > 300
OR molecule has cLogP > 3
THEN molecule is not lead-like
If you dive into the syntax of computer programming languages, you’ll see different ways that they implement these logical statements. In Manifold, we’ve tried to boil this complex syntax down to 3 easy-to-use filters which closely match the desired use-cases for chemists.
Structure filters: place a strict requirement for or exclusion of a structure.
- This can be done using one of the pre-defined structures (including protecting groups like NFmoc, NBoc, and NCbz) or you can even draw your own.
Numeric range filters: for initial screening of compounds.
- These include cLogP, MW, and number of heavy atoms.
Alerts: exclude results which trigger medicinal chemistry alerts.
- Choose a set of medicinal chemistry alerts from industry leaders such as Novartis, PAINS, GSK, and others.
These types of detailed filtering queries which used to end in days of back and forth emails, hand filtering, or utter frustration, can now be handled in mere seconds.
Illustrative Examples
Let’s dive into a couple examples to get a feel for what you can expect out of the tool.DEL & Scaffold Hopping
Boolean Logic filtering is especially useful to help propose scaffold hops based on knowledge of the available purchasable scaffolds, especially in their protected forms.Let’s imagine that the spirocyclic scaffold of Figure 2a comes up as an enriched diamine scaffold from a DEL screen. In a scaffold hopping exploration, you are interested in looking for similar scaffolds that may not have been in the DEL library.
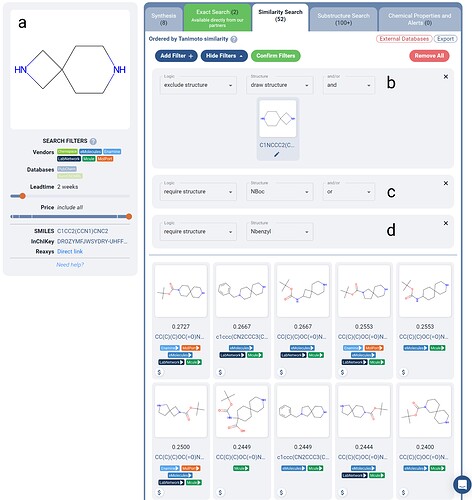
Figure 2. Boolean Logic-filtered similarity search for a spirocyclic scaffold (a) from a DEL screen. The scaffold itself is excluded (b) to yield different scaffolds only, and either an NBoc (c) or Nbenzyl (d) protecting group is required in each result. Over 50 in-stock purchasable molecules are returned, where the top results contain distinct spirocycles with the potential for use in scaffold hopping.
To only see new scaffolds, we can apply an exclude structure filter and draw the original scaffold (Figure 2b). You could imagine adding in other excludes here as well if there were other similar scaffolds you want to rule out. We also applied two require structure filters, NBoc and Nbenzyl protecting groups (Figure 2c-d). Notice the OR between these two filters - this allows you to require that one or the other is present, allowing more versatility in the results returned.
The results returned in Figure 2 have been filtered with the vendor filters on the left to exclude PubChem and SureChEMBL databases, to narrow in on purchasable scaffolds. Lead time is set to 2 weeks to see in-stock results only. With these additional constraints, there are over 50 results, ranked via similarity to the original scaffold.
Of course, these similarity values are quite low, but this is due to the requirement of the protecting group which is not present in the original scaffold. Only the top 10 results are shown in Figure 2 for brevity, where 6 of the 10 are unique spirocyclic scaffolds, and 4 of the 10 are diamines.
We tried to get comparable results from other similarity search engines for this DEL scaffold, with varied spirocyclic scaffolds in their protected form. Unfortunately, the only way we have found to (partially) accomplish this is by querying the scaffold itself in its NBoc or Nbenzyl protected form.
When it comes to fast and simple retrieval of these analogs for assessment of potential for scaffold hopping, you might consider leveraging Manifold with Boolean Logic filters.
PROTAC Linkers
Coinciding with the recent release of the structures of the first Phase 2 clinical proteolysis targeting chimeras (PROTACs) protein degraders, our next example will focus on PROTAC linkers.For an overview of how PROTACs work, we’d like to point the reader to a great summary put together by XVIVO and Arvinas. In short, a PROTAC moiety is composed of two ligands (target protein & E3 ligase binders) joined by a linker, which allows for controlled ubiquitination of the target protein for protein degradation.
The linker composition and structure has been shown to significantly affect the physicochemical properties of the PROTAC moiety, and in turn, potency. Thus, optimization of the linker chemistry is a rapidly developing frontier in the field.
A recent PROTAC work by Shilpi Arora and coworkers at X-Chem uses DEL screening to identify novel protein binding features via determining the linker positioning and binding elements from the screen. The resulting protein degraders were shown to inhibit tumor growth in a mouse model for breast cancer. Interestingly, Arora and team carried out rapid and systematic PROTAC synthesis utilizing the Click Chemistry toolkit from researchers at Amgen.
The click-chemistry utilized for systematic PROTAC synthesis works by clicking together ligase inhibitors with target protein ligands to rapidly craft PROTAC libraries. Let’s use Manifold and Boolean Logic filters to see how we can further streamline this methodology through direct purchase of variable alkyne linkers to be utilized in the chemistry.
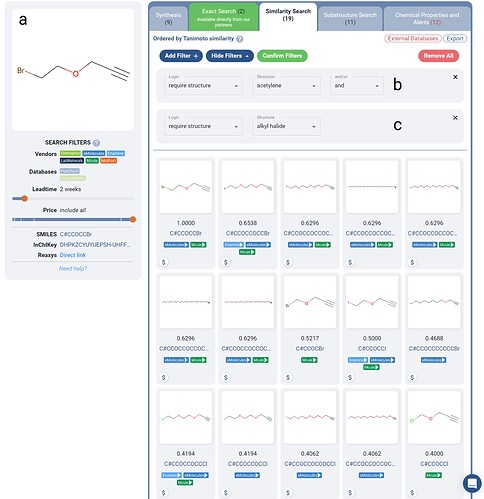
Figure 3. A filtered search for a set of purchasable alkynes for use in PROTAC click-chemistry similar to the queried alkyne (a). Boolean Logic filters are applied such that the molecules returned are required to contain an alkyne(b) and an alkyl-halide group(c). 19 in-stock purchasable molecules are returned with variable length chains, with an alkyne on one end and an alkyl-halide on the other.
A common intermediate for formation of the click-chemistry linkers in the toolkit are chains with alkynes with alkyl halides on either side. Using Manifold, we crafted a similarity search for one such compound (Figure 3), where we used Boolean Logic filters to require the presence of an alkyne and an alkyl halide in similar molecules (Figure 3b-c).
As shown in Figure 3, the above filters yield 19 purchasable in-stock (2 week lead time) molecules which could be utilized in PROTAC click chemistry. Search strategies like this could be utilized to help accelerate building out PROTAC libraries which systematically investigate the linker composition and structure.
Fragment Libraries
For this last example, we’ll use the recent work by K. Marks and coworkers, an interesting fragment-to-clinical candidate campaign targeting MAT2A Inhibitors (highlighted recently on the Practical Fragments blog), where the initial fragment core is intact in the clinical compound, to illustrate an example of building out a fragment library with Manifold + Boolean Logic filters.In this work, an initial screen of over 2000 fragments confirmed the hit illustrated in Figure 4a. Next the researchers performed a virtual hit expansion similarity search with constraints of Tanimoto >80%; 200 < MW < 350, which yielded them 54 purchasable compounds. Tests for enzymatic inhibition of MAT2A and SPR binding yielded a most-potent MAT2A inhibitor, which we are displaying in Figure 4a.
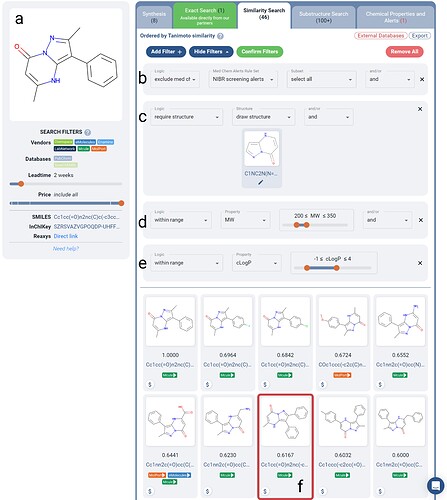
Figure 4. Search for purchasable similar fragments to the initial fragment screen hit (a) as described in the referenced work in the main text. Hits must not trigger any Novartis screening medicinal chemistry alert (b), are required to contain the original scaffold (c), and must lie within ranges: 200 < MW < 350 (d) and -1 < cLogP < 4 (e). The most potent inhibitor that the researchers found is returned within the top 10 hits on Manifold (f).
We repeated their similarity search using Manifold (Figure 4b-e). Addition of Boolean Logic filters allows a chemist in a fast no-code manner, to narrow in on purchasable similar compounds with desired constraints. As the researchers have done, we applied a molecular weight constraint, as well as cLogP numeric range constraint. We also require explicitly that the fragment scaffold is maintained, and that any hit which contains a Novartis screening alert is excluded. This extent of search fine-tuning is not available via other interactive search tools, and previously only available via working with cheminformatics specialists.
As shown in Figure 4, once further filtered to compounds available in 2 weeks, we can see that 46 similar compounds are found with the desired constraints; narrowed down 13% from the set that the researchers worked with. Interestingly, the eventual most-potent inhibitor sits at number 8 in Manifold’s results (as ranked via similarity to the original fragment).
Although many think of similarity and substructure searching as solved problems, there is much room to make sophisticated search queries accessible. We really hope these examples illustrate interesting use cases that are facilitated by Manifold’s Boolean Logic filters.