Dear COVID Moonshot Community,
We are happy to release results from our first round of screening. The data so far includes 28 hits with over 50% inhibition at 50 μm, 8 hits with IC50 <10 μM, and over 20 new crystal structures of compounds bound to the main protease. Since you’ve made it to this post from Twitter or our email, I’ll assume you want more details!
1. Testing for main protease activity?
First, a big thank you to @londonir and Haim Barr at Weizmann who worked so hard to get this assay up and running.
We just got new results in today, and even more IC50 values will be back by Tuesday. All results (including Trypsin counterscreen and solubility data obtained at Enamine) can be viewed on the PostEra website here: https://covid.postera.ai/covid/activity_data
Procedure:
Compounds were seeded into assay-ready plates (Greiner 384 low volume 784900) using an Echo 555 acoustic dispenser, and DMSO was back-filled for a uniform concentration in assay plates (maximum 1%). Screening assays were performed in duplicate at 20 µM and 50 µM. Hits of greater than 50% inhibition at 50 µM were confirmed by dose response assays. Reagents for Mpro assay reagents were dispensed into the assay plate in 10 µl volumes for a final of 20 µl. Final reaction concentrations were 20 mM HEPES pH=7.3, 1mM TCEP, 50 mM NaCl, 0.01% Tween-20, 10% glycerol, 5 nM Mpro, 375 nM fluorogenic peptide substrate ([5-FAM]-AVLQSGFR-[Lys(Dabcyl)]-K-amide). Mpro was pre-incubated for 15 minutes at room temperature with compound before addition of substrate. Protease reaction was measured continuously in a BMG Pherastar FS with a 480/520 ex/em filter set. Data was mapped and normalized in Genedata Screener prior to loading to CDD vault.
Overall, we have been pleasantly surprised by the reproducibility of the assays. Here are two examples:
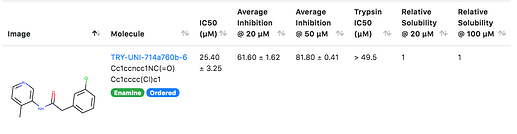
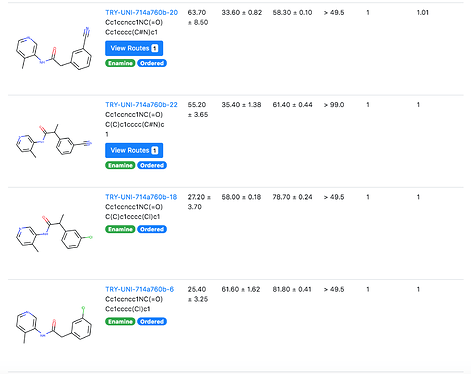
TRY-UNI-714a760b-6
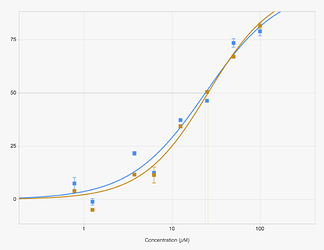
which has two IC50 curves shown here, which agree quite well.
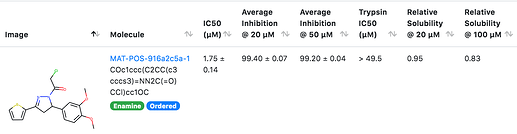
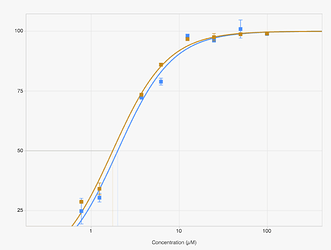
again has dose-response curves that agree nicely.
In terms of testing, we also plan to perform thiol reactivity assays in the next few days for the ~80 covalent compounds tested so far.
- Getting Structures
A huge thank you to @frankvondelft, @Daren_Fearon, @AnthonyA, @Waztom , @reskyner and the whole Diamond UK team who has been working on getting these amazing structures. The speed at which this team has obtained over 20 new structures beyond the initial fragments is incredible. You can browse the structures on Fragalysis or download all structures from the top of the page as shown on the right of the screenshot below
We have also collected many other structures from the literature which may be of use in designs here: https://covid.postera.ai/covid/structures#literature_SARS-COV-2
3. What’s Next?
We are now in the process of synthesizing and assaying another 200 compounds with plans to have hundreds of more follow up compounds to expand and optimize the hits that we have obtained. As a recap, we’ve received over 4600 unique designs and selected 700 compounds to make and test. In the first wave we focused on compounds that could be rapidly synthesized. We rapidly triaged compounds for synthetic feasibility and sent our first batch of compounds with synthetic route designs at the start of April (PostEra), and have successfully completed synthesis (Enamine), main protease biochemical assays (Weizmann Institute) and X-ray crystallography (Diamond/Oxford).
From you all, we hope to continue obtaining insights and designs as we continue to look for promising leads. Expanding upon the current actives is great, and @frankvondelft has even set up a category seeking suggestions on how to best merge and enumerate the current fragments with structures. We are also setting up the infrastructure to better gain insight from community simulation results.
We now have over 20 synthetic chemists working on the project between the great folks at Sai Life Sciences and Enamine. Let’s make some stuff!
And for those of you who want to pay for your own designs to be synthesized and/or tested, please see this post (and make sure to respond to the post if your are sending compounds for synthesis/testing).
Some other highlights
I haven’t actually gotten a ton of time to look at the data (beyond checking it is in the right format). My week has been filled with mostly data processing, logistics, and some web development. But I’ll just point out a few things that I think may be of interest.
- Covalent compounds: A lot of the early hits are chloroacetamides or Ugi compounds with an acrylamide warhead. Some work needs to be done to disentangle K_i vs K_{inact}. @RGlen, @BAL, and folks at Boehringer Ingelheim are doing some work to help probe this problem computationally!
-
3-amino pyridine non-covalent hits Some of the med-chemists who saw the data first, noted that the ligand efficiency of this series of non-competitive inhibitors is quite good for a challenging protein like this.
-
MAT-POS-bfefc3ea-3, which is Z LVG CHN2, a cysteine protease inhibitor from the ReFrame library tested in https://www.biorxiv.org/content/10.1101/2020.04.16.044016v1.full.pdf, did not show great activity in our assays (10.4 ± 2.36 % inhibition at 50 μm). However, it still showed promising results against the virus in the original paper. We thought it was worth testing if it was binding to MPro, but I guess we’ll just let it remain a mystery of biology
 .
.
Questions
I’m sure many of you have questions. Please reply to this thread if so, We’ll try to respond, though sending this out near midnight my time (PST) is perhaps a terrible idea!
Thanks
And lastly, thanks to all of you in the community who continue to make this initiative what it is.
-Matt, and the rest of the PostEra team helping out on this project (@alphalee, @aaron.morris, @ajajack, @milan.cvitkovic)

 :
: