I thought I’d make a quick dataset that could be handy for someone maybe.
@reskyner right at the start as a proof of concept for site labelling in Fragalysis made a file with bromide atoms marking sites of interest in the active site —I used that file (ref_atoms3.pdb) and the hit determined positions of the submissions to map where the submission lie.
- HTML with pictures: https://www.well.ox.ac.uk/~matteo/submissions_site_distances.html
- Excel without pictures: locations.xlsx (509.6 KB)
The code used is just:
compounds = Chem.SDMolSupplier('Fragmenstein_permissive_rescored_20200610.sdf')
# Retrieve the positions
ref = Chem.MolFromPDBFile('ref_atoms3.pdb')
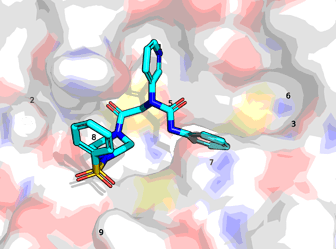
ref_names = {'3': 'P5-sidechain', '6': 'P4-sidechain', '7': 'P2-sidechain', '1': 'P1-sidechain', '5': 'P1-backbone',
'8': 'P1′-backbone', '2': 'P2′-sidechain', '9': 'P4′-sidechain'}
ref_conf = ref.GetConformer()
ref_points = dict(zip([ref_names[str(a.GetPDBResidueInfo().GetResidueNumber())] for a in ref.GetAtoms()],
[ref_conf.GetAtomPosition(i) for i in range(ref.GetNumAtoms())]))
# ref_points is a dictionary with a nice name key and Point3D value
# get all distances
data = []
for mol in compounds:
conf = mol.GetConformer()
d = {'Name': mol.GetProp('_Name'), 'SMILES': mol.GetProp('smiles')}
for k, v in ref_points.items():
ds = [conf.GetAtomPosition(i).Distance(v) for i in range(mol.GetNumAtoms())]
d[k] = min(ds)
data.append(d)
distances = pd.DataFrame(data)[['Name', 'SMILES']+list(ref_names.values())].round(1).iloc[1:]
PandasTools.AddMoleculeColumnToFrame(distances,'SMILES','Molecule',includeFingerprints=False)
The covalents have four combinations of warheads (‘Acrylamide’, ‘Chloroacetamide’, ‘Vinylsulfonamide’, ‘Nitrile’ —‘Bromoalkyne’ and ‘Aurothiol’ warheads disabled), but that should make little difference, so just ignore the suffix. The SMILES are already covalent (with *), sorry.