Topic automatically created for discussing the designs at:
https://covid.postera.ai/covid/submissions/PET-UNK-c9c1e0d8
Thanks @pwkenny,
I’ll note that JAN-GHE-83b26c96-12 and TRY-UNI-714a760b-6 were already assayed and showed very little difference in IC50 from the Cl --> Br change
Would you expect any difffernce due to the pyridine --> isoquinoline change?
Thanks, Matt, I would anticipate similar [Cl->Br] differences for the pyridine and isoquinoline and I would suggest not synthesizing the bromophenyl analog for the isoquinoline at this stage. I had included it as something that needed to be checked and the results for pyridine (of which I’d been unaware) provide the necessary information.
Hi Matt, I see that the PET-UNK-c9c1e0d8-4 has been ordered while PET-UNK-c9c1e0d8-3 has not. Could I ask why the 5-membered ring is preferred to 6-membered ring?
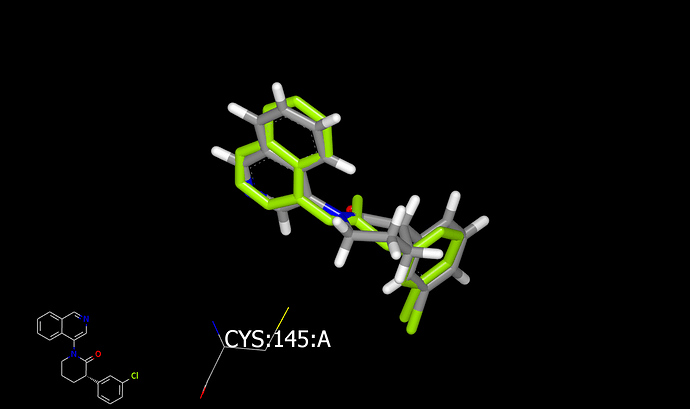
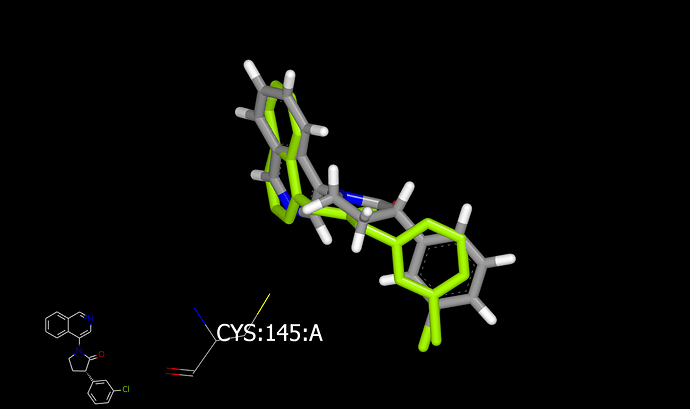
Here are a couple of graphics showing dockings (into x10959) for the 6-membered and 5-membered rings:
There are two reasons that I would favor the 6-membered ring over the 5-membered ring. First, the 6-membered ring appears fit more easily into the binding site (and is a better mimic of the x10959 ligand. Second, the axial hydrogen of the 6-membered ring is directed at catalytic cysteine. Substituents on carbon next to amide nitrogen in saturated rings typically favor an axial orientation making the axial CH the attachment vector.
I’ll write up some notes (including analysis of CSD) over the next couple of weeks although I thought that it’d be a good idea to share these thoughts now. One the methylenes of the 6-membered ring can be replaced with oxygen [piperidone => morpholone) without changing the molecular shape or attachment vector orientations. I will submit this as a design later this week since the ring system may be more amenable to synthesis.
Hi Matt, just letting you know that the [4-CH2=>-4-O] modification of PET-UNK-c9c1e0d8-3 has now been submitted as PET-UNK-431b3bfb
Hi Matt @mc-robinson, looks like the PET-UNK-c9c1e0d8-4 shows comparable potency to the ADA-UCB-6c2cb422-1 parent structure. I would recommend synthesizing PET-UNK-c9c1e0d8-3, which appears to fit more easily into the binding site and is also likely to provide better access to the catalytic cysteine (e.g. PET-UNK-3e354a91-1). The design rationale for these two cyclic analogs of ADA-UCB-6c2cb422-1 is that they present additional vectors for synthetic elaboration (although any potency gains are clearly welcome) and I’ll mention @edgriffen since we were discussing his EDJ-MED-28ec730d submisssion in the context of access to the S1’ subsite. A crystal structure would provide information about the vector orientations and I’ll also mention @frankvondelft.
Thanks @pwkenny, we are a little backed up in terms of synthesis because we are synthesizing compounds for PK, but I’ll keep that one in the queue.