Topic automatically created for discussing the designs at:
https://covid.postera.ai/covid/submissions/PET-UNK-4880b143
The PET-UNK-4880b143-1 design merges the isoquinolinyl amide PET-UNK-29afea89-2 with the fragment AAR-POS-d2a4d1df-1 that had been identified (x0072) in the crystallographic screen. The x12207 crystal structure (ligand: EDJ-MED-e4b030d8-13 ) was used for modeling and I’ll mention @frankvondelft in case there is interest in how crystal structures (and fragment hits) are being used in design. I’m assuming that the sulfone would be accessible via Michael addition of EDG-MED-971238d3-1 (or its precursor) to methylvinylsulfone. I would anticipate that this structural elaboration will extend to structural variations of the chromane ring.
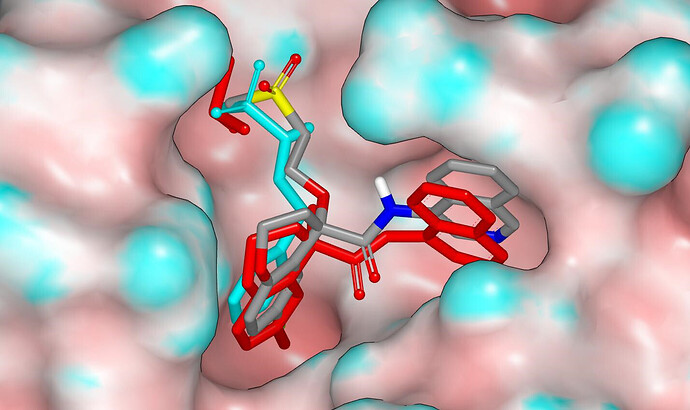
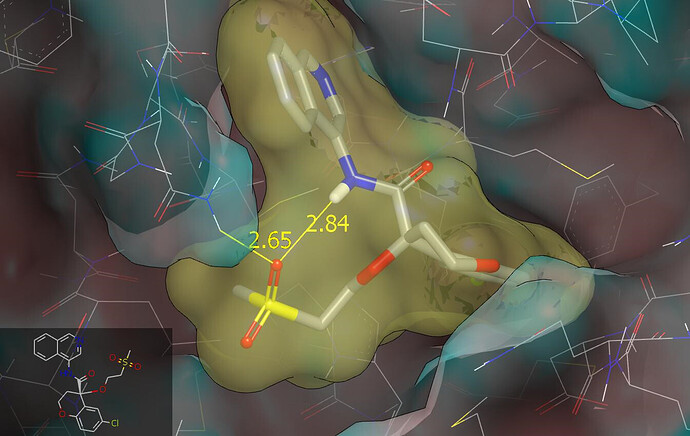
The binding mode was modelled using Szybki (OpenEye) and energy was minimized using the MMFF94S force field. The isoquinolinyl amide substructure flattens somewhat in the course of energy minimization and I’ve manually returned it to what is observed in crystal structures. I rotated (by 120°) the methylsulfonyl group of the AAR-POS-d2a4d1df-1 fragment around the S-N bond to set the dihedral to be consistent with what is observed for N-alkylsulfonamides in the CSD (I’m assuming that carbon and oxygen atoms cannot be distinguished in the protein structure). The PET-UNK-4880b143-1 design has equivalent connectivity (two atom type differences between the chains linking methylsulfonyl to chromane ring) to the DAR-DIA-6a508060-13 design and I’ll mention @Daren_Fearon . The graphic below shows the molecular surface of the protein (colored by curvature), the PET-UNK-4880b143-1 design (colored by atom type), the ligand and a relevant DMSO molecule from the x12207 crystal structure (red) and the fragment from the x0072 crystal structure (cyan). The graphic was generated from the pdb file associated with this submission.
Thanks @pwkenny , good spot. Any reason you wouldn’t want to use the sulfonamide straight from the x0072?
The sulfonamide won’t work with the oxygen-based linker because there’ll be two heteroatoms on an acyclic methylene (concerns about stability) although the carbon-based linker (e.g. DAR-DIA-6a508060-13) could allow a sulfonamide to be positioned in an appropriate manner. The sulfonamide NH of the fragment AAR-POS-d2a4d1df-1 doesn’t appear to be interacting with the protein and it can probably be replaced with CH2. I have more confidence in the oxygen-based linker than the carbon-based linker to position the methylsulfonyl group in an appropriate manner.
I would anticipate ADME characteristics to be better for the sulfone than for the sulfonamide since the oxygens of the former are likely to be better hydrogen bond acceptors than the the oxygens of the latter. Also the presence of hydrogen bond donors can adversely affect solubility (@edgriffen may have data that allows this question to be addressed).As an aromatic substituent, the methylsulfonyl group is seen as beneficial from the ADME perspective. That said, DAR-DIA-6a508060-13 also looks like an attractive candidate for synthesis.
I submitted DAR-DIA-6a508060-13 before we had the structure for EDG-MED-971238d3-5 - comparing the fragment with the methoxy and methylamine compounds I think Pete has a point about the oxygen-based vs carbon based linker and positioning of the methyl sulfonyl group although who knows how it might rotate with something bulkier.
The oxygen based linker tends to direct the substituent away from the amide NH and my concern about the carbon-based linker is that it might direct the substituent toward the amide NH. That said, I don’t currently have any evidence that this would be the case and I think DAR-DIA-6a508060-13 looks to be worth synthesizing. I recommend re-submitting it as a single enantiomer along with the N-methyl analog (the sulfonamide NH doesn’t appear to be doing anything and capping it may be beneficial the ADME).
I’ve been taking a closer look at the carbon-based linker for the sulfonamide is not currently looking good. The oxygen-based linker can orient so that the oxygen-carbon bond is directed away from the amide NH and the carbon-based linker doesn’t look like it can easily get into this conformation. I’m still checking things but I thought that I’d let you know since I’d previously suggested re-submitting DAR-DIA-6a508060-13 as a single enantiomer.
Thanks for the heads up Pete.
Hi Daren,
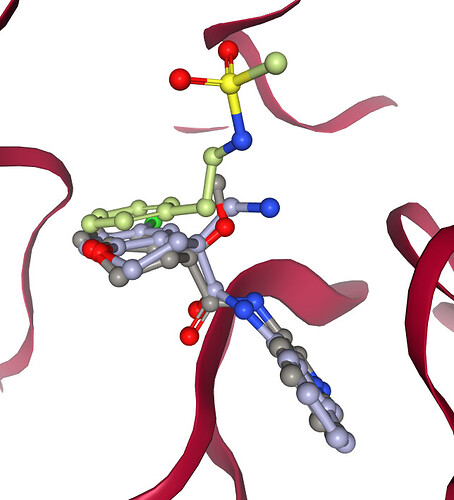
I’ll do some more extensive checks since I want to be sure (or at least as sure as one can be from modeling). One problem with cyclic frameworks like the chromane (cyclic link between the P2 phenyl and the benzylic carbon) is that they don’t appear to provide good access to other regions in the binding site. I’d anticipate that making a cyclic link between the amide nitrogen and the benzylic carbon of the P2 substituent will provide better access to the S1’ subsite without cutting off access to the S4 subsite. Here’s a graphic, which might also be of interest to @frankvondelft, showing the sulfonamide fragment AAR-POS-d2a4d1df-1 (with S-N torsion set for consistency with what is observed in CSD) from x0072 with the A-chain protein molecular surface (colored by curvature) and ligand (PET-UNK-c9c1e0d8-4) from n0066. The graphic also points to potential benefits of N-methylation of the sulfonamide.
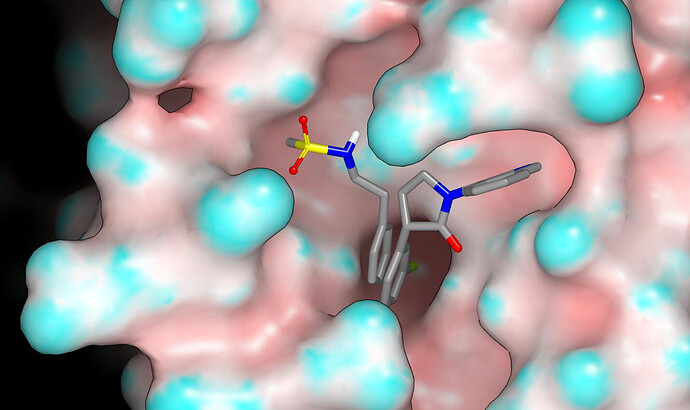
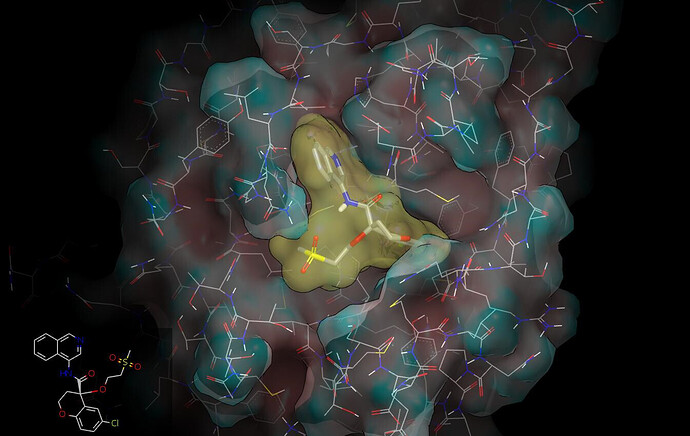
I have used the P0157 A-chain crystal structure to update the proposed binding mode: PET-UNK-4880b143-1_updated_bindng_mode.pdb (466.6 KB). Here are couple graphics to illustrate the new binding mode: