Topic automatically created for discussing the designs at:
https://covid.postera.ai/covid/submissions/PET-UNK-3bb57da2
I’ve used the crystal structure (P0157) that was recently determined for the MPro complex with PET-UNK-29afea89-2 to generate some more designs. I’ll mention @Daren_Fearon @frankvondelft since they may be interested in how the crystal structures are being used and because of their interest in the PET-UNK-4880b143-1 design. The pdb file associated with the submission includes updates for binding modes proposed for the designs in the PET-UNK-824b5c6a submission.
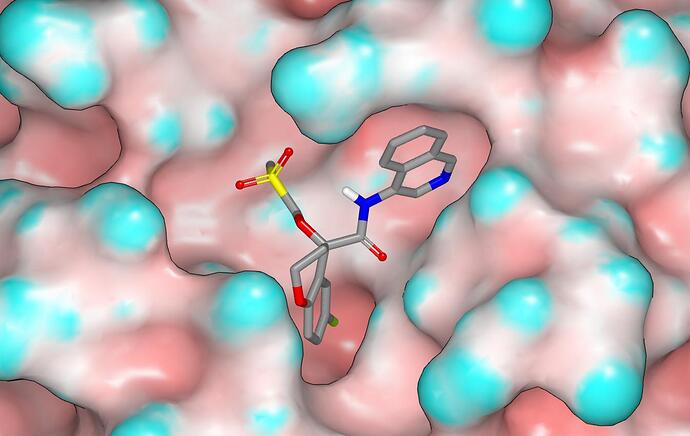
The sulfone PET-UNK-3bb57da2-1 can make hydrophobic contact with the protein surface that is equivalent to the n-propyl analog PET-UNK-824b5c6a-2 (it may be worth synthesizing this design in order to assess the value of the hydrophobic contact since it is likely to be more readily accessible) while directing the polarity of the sulfonyl group away from the protein surface. The N,N-dimethyl sulfonamide PET-UNK-3bb57da2-2 appears to make even better hydrophobic contact with the protein surface. I do have a concern about the 180 degree O-C-S-[C,N] dihedral in the binding modes proposed for these two designs although there are very few representative structures in the CSD and I don’t have access to the QM software that I would typically use to address issues like these.
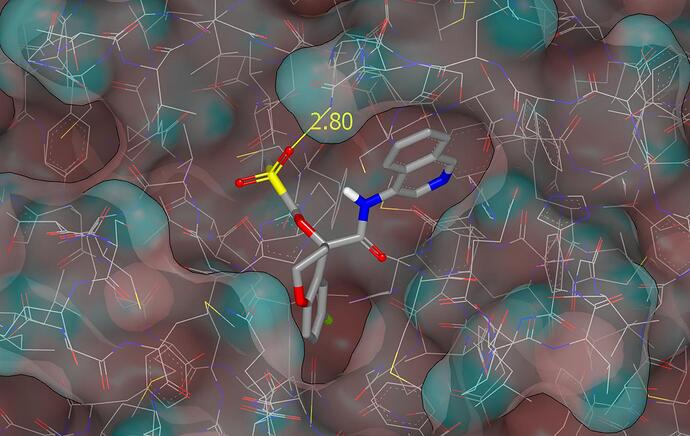
While setting up the first design, I noticed that one of the sulfonyl oxygens was close to the amide oxygen of the N142 and the amide has been rotated by 180 degrees for modeling. Here are a couple of graphics (molecular surface is colored by curvature) showing the binding mode proposed for PET-UNK-3bb57da2-1 with one of the sulfonyl oxygens accepting a hydrogen bond from the N142 side chain.
Thanks @pwkenny . Btw, there is a way to upload 3D models to Fragalysis - it’s necessarily faffy to get the format right, but nothing a geek can’t sort out - instructions are here on the forum, posted by Rachael Skyner sometime last summer. (The features weren’t working well since then, but all bugs are now finally flushed out, as far as we can tell.)
Hi Frank, I’ll take a look and it’s generally easy to write the structure files in different formats with OpenEye’s VIDA software.