Topic automatically created for discussing the designs at:
https://covid.postera.ai/covid/submissions/PET-UNK-1e13ef09
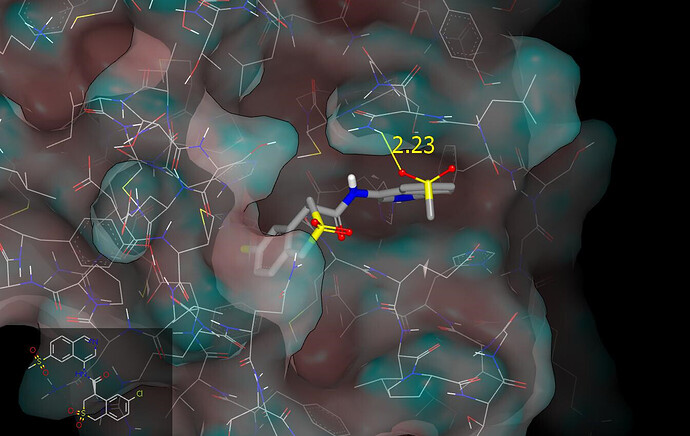
It looks like a 6-methanesulfonyl substituent on isoquinoline can accept a hydrogen bond from the protein (side chain amide of N142) and I’ll mention @edgriffen @mc-robinson @alphalee @Ben_DNDi in case it’s of interest. Here is a graphic (protein surface colored by curvature) illustrating the binding mode predicted for the cyclic sulfone PET-UNK-1e13ef09-2 (it’s the R enantiomer of the previously submitted ALP-POS-e6e0c683-4 racemate and has been labelled as such the PDB file associated with this submission).
So the question is Pete, when we look at the 6-MeSO2 vs the 6-H for matched pairs the 6-MeSO2 looks consistently poor value, I buy the argument that it should be contacting N142 - but that doesn’t seem to be doing much good if it is:
Hi Ed, the 6-methylsulfonyl substitution certainly looks underwhelming from a potency perspective and I presume that it brings some concrete benefits that compensate for the hit in potency. Methylsulfonyl is electron-withdrawing and C6 in the ‘other’ ring ‘communicates’ most effectively with the isoquinoline nitrogen (see submission notes for PET-UNK-03fd2068). The side chain amide of N142 is solvent exposed and I wouldn’t expect the hydrogen bond to contribute much to affinity even if it does form (it was just something that I noticed in the energy-minimized complex and shared in case of interest). The 6-cyanoisoquinoline BEN-DND-c852c98b-1 is about threefold less potent than its 6-methylsulfonyl analog BEN-DND-c852c98b-5 which is interesting because the nitrile should be more easily accommodated (the methylsulfonyl group is bulkier than cyano and less able to present its polarity to solvent in the manner that nitrile can).
If faced with a potential metabolism problem with isoquinoline, my opening move would be actually be F at C7 (F has minimal steric footprint while C6 ‘feels’ the electron-withdrawing effect of the isoquinoline to a greater extent than C7 and is therefore less in need of protection). One interesting design that I noticed recently is @ruby’s RUB-POS-1325a9ea-14. The des-methyl analog RUB-POS-1325a9ea-12 (pIC50 = 5.7) is more potent than either of the naphthyridine analogs MAT-POS-f7918075-5(pIC50 = 5.4) and MAT-POS-bb423b95-1 (pIC50 = 5.1). My view is that the hydrogen bond donor of the pyrazolopyridne will not be beneficial for affinity and I’d anticipate that capping it with methyl will result in increased potency.
The central challenge right now is rodent metabolism where the isoquinoline looks like the culprit (less so in human, dog to be tested), the 6-MeSO2 is one of the few groups that improves metabolism a bit, but not enough, so the search is on for something better - or an IQ replacement. The 7-F, and 7-Cl are both in for synthesis.
Hi Ed,
As far as I can see the 6-methylsulfonyl isoquinolines that have been made are all racemic. Does this mean that the metabolic stability studies have been carried out using racemic material? Also have you assessed plasma stability?
As noted earlier in the discussion, I consider 1-methylpyrazolopyridine (as in RUB-POS-1325a9ea-14 ) to be a potentially interesting isoquinoline replacement. I see N1 as ‘feeding’ the lone pair of the nitrogen in the 6-membered ring while N2 provides the 5-membered ring with a degree of protection from metabolism. According to the system, the status of RUB-POS-1325a9ea-14 is ‘synthesis in progress’ and I would recommend chasing this up.
Another tactic that I would recommend is to add methoxy next to the nitrogen in the ‘other’ ring of each naphthyridine as exemplified by PET-UNK-f4e47ebd-1 and PET-UNK-f4e47ebd-2. As discussed in the PET-UNK-f4e47ebd submission notes, an adjacent methoxy substituent would be expected to reduce the desolvation penalty for the nitrogen (hopefully reducing the hit in potency associated with aza-substitution of isoquinoline). The methoxy substituent would also tend to ‘feed’ the lone pair of the nitrogen that accepts a hydrogen bond from the protein. I think that this would work better with 6-methoxy and 7-aza than vice versa although I would recommend evaluating both options (the initial question of whether the methoxy substitution reduces the hit in potency would be most easily addressed by synthesizing the 3-chlorophenylacetamides PET-UNK-f4e47ebd-1 and PET-UNK-f4e47ebd-2).