Topic automatically created for discussing the designs at:
https://covid.postera.ai/covid/submissions/9269dfbd-0386-44e3-b0e3-617ca9cece99
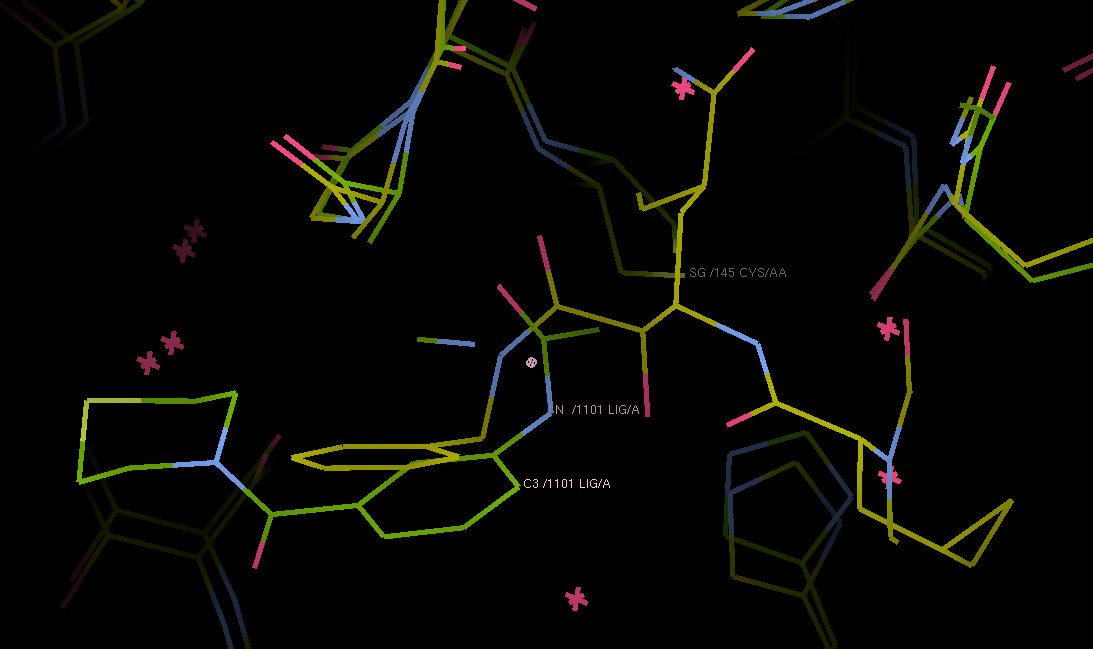
Detail of the overlay of the ketoamide inhibitor in PDB ID: 6y2g (yellow) and fragment 0786 (green). I think the active site on the left is sufficiently flexible to merge them and generate a fragment-based ketoamide inhbitor.
P.S. Cys 145 is dimly visible in the back, for reference
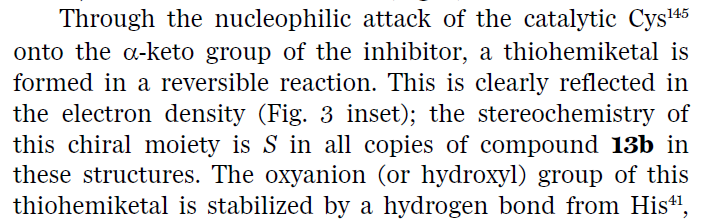
Hi Joost, I was wondering how the covalent inhibition using alpha-ketoamides is working in this situation. The thiohemiacetal, which is formed after addition of the cystein residue will be very unstable and the initial bond formation will be easily reversible. Unless there is some kind of specific stabilisation of the alkoxide from residues around? Or am I missing something and is there a subsequent irreversible reaction or is it meant to be a reversible inhibitor?
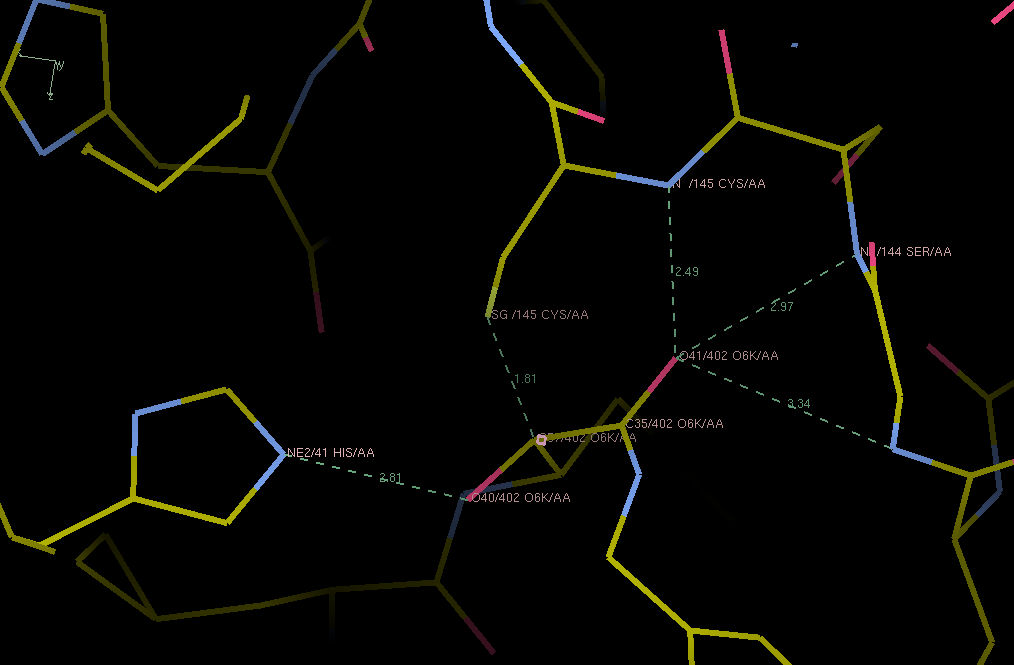
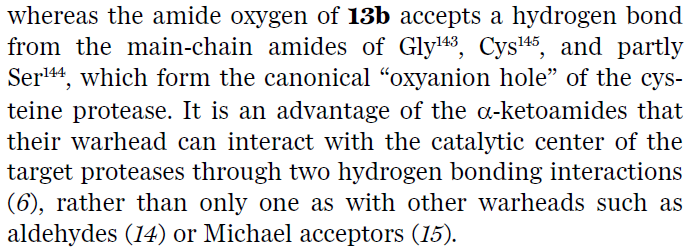
Hi Jan. Thanks for the question. The authors say the bond is reversible, so I guess that means it is not really extremely tight. They describe that both keto-moieties are bound in their own respective anion holes (see screenshot). One to a histidine, the other to a series of backbone nitrogens, so a nice double interactions. Makes you wonder what is the ‘real’ anion hole in the protease. According to Zhang et al in the Science paper, this is the site of the backbone interactions. Nicely, this is also where all of the chloroacetamide oxygens bind, so that would make sense. Anyway, the ketoamide inhibitor only has uM affinity, to there is a lot to be improved - best Joost
Thank you for the explanation. Very interesting.