Topic automatically created for discussing the designs at:
https://covid.postera.ai/covid/submissions/FRA-DIA-a1f3a927
Modelling by scribble, this is what it would look like:
and here’s the Fragalysis scene: https://fragalysis.diamond.ac.uk/viewer/react/projects/303/243 (it might have gone off when you view it, known bug, grrr) or simply the direct link to the structure: https://fragalysis.diamond.ac.uk/viewer/react/preview/direct/target/Mpro/mols/VLA-UCB-1dbca3b4-15/L/P/C
According to Manifold, it would be easy to make (if I understand the output correctly): https://postera.ai/manifold/45f5d015-02c1-4899-b980-73a81ca80100/0/
Hi Frank @frankvondelft this design is analogous to BEN-DND-61647d40-20 from @Ben_DNDi and NIR-WEI-75ed5c39-1 from @londonir. I think the idea would be better tested by synthesizing BEN-DND-61647d40-20 or its isoquinoline analog. A covalent warhead (nitrile) can be linked directly (using cyanogen bromide) to the amide nitrogen as in the PET-UNK-5ecb6237-1 design although I think that PET-UNK-a692de38-1 would have a better chance hitting the cysteine. Both nitrile designs address the problem that substitution of the amide nitrogen with sp3 carbon is expected to invert the cis/trans geometric preference of the amide (see PET-UNK-bbe8d7ff-2). The primary rationale for the PET-UNK-c9c1e0d8-3 design is to enable a warhead to be presented to the catalytic cysteine (e.g. PET-UNK-3e354a91-1).
Agree this sort of design is one not to be missed, could be really interesting. Either the optimized version from Frank or the very simplified version from my original submission, or a hybridized version of the two - whichever is easiest to make I guess?
B
The benzofuran seems to matter, so it’s not that analogous to BEN-DND-61647d40-20.
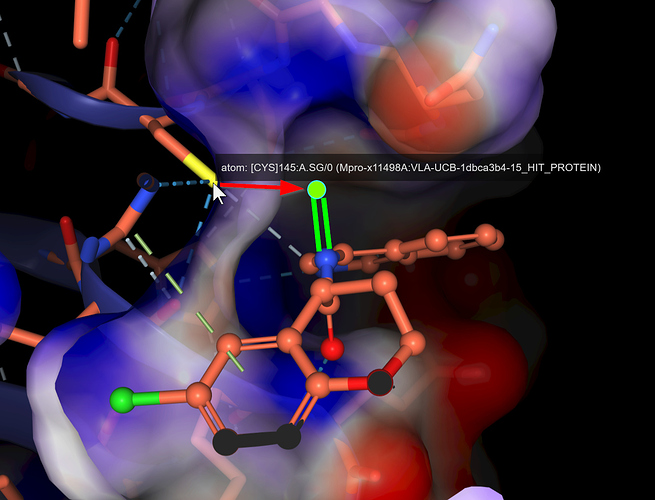
@pwkenny, eyballing x11498, the electrophile would be pretty close to Cys145, unless I misunderstand the reaction: what I think what might be needed is something in P2 to give His41 some help in deprotonating the SH group, the way the natural substrate does (again, speaking loosely from memory).
@pwkenny can you spell out what the cis/trans inversion will entail? The amide nitrogen doesn’t interact with the protein as far as I can tell (x11498), unless it’s through a water (apologies, a Fragalysis bug is hiding them) - I missing something?
Hi Ben
My view is that targeting the cysteine needs to be done in a systematic manner and carefully coordinated. The difference between BEN-DND-61647d40-20 and FRA-DIA-a1f3a927-1 is that the former is closely related to the known inhibitor EDJ-MED-49816e9b-1 while I don’t believe that anything like the parent (>CH2 rather than >C=CH2) ketone of FRA-DIA-a1f3a927-1 has been assayed (or even synthesized). It’s also possible that the isoquinoline may not confer the same benefits when it is linked by carbon (as opposed to amide nitrogen) and I’ve submitted the PET-UNK-8922bd3c-1 design to test this hyopthesis.
Hi Frank,
My main concern with FRA-DIA-a1f3a927-1 is that the parent structure (a ketone with methylene linking the carbonyl to the isoquinoline) has not been shown to be an inhibitor. I would expect the 3-chlorobenzyl group of BEN-DND-61647d40-20 to occupy S2 and believe that this design (or the analogous design derived from EDJ-MED-49816e9b-1) could be used to explore the possibility of covalent bond formation with the catalytic cysteine. The Michael acceptor could be made more electrophilic by varying the heterocycle although I’d expect this to destabilize the non-covalently bound complex. I see the first step in targeting the catalytic cysteine as establishing that the covalent bond actually forms and the exploration can conducted using structurally prototypical compounds.
The inversion of cis/trans preference is a separate issue. While the amide NH might look like a great vector for exploitation, linking sp3 carbon to the amide nitrogen would be expected to ‘flip’ the amide as discussed in these notes. This effect is reflected in the assay results for MAT-POS-bb423b95-7 which is 1.7 log units less potent than ADA-UCB-6c2cb422-1 in the fluorescence assay. Attaching sp carbon ( ADA-UCB-6c2cb422-1) or nitrogen (PET-UNK-a692de38-1) to the amide nitrogen would be expected to ‘preserve’ the trans amide geometry required for targeting the catalytic cysteine.
Thanks Peter - you seem to be discussing your design, not mine; I am proposing to replace the N with an sp2 carbon, and lose the amide bond.
That said: my chemistry abandons me: does that thing tautomerise spontaneously, or does one need to worry about which isomer is formed?
Hi Frank,
I think that FRA-DIA-a1f3a927-1 will exist predominantly as the tautomer as drawn. However, I’d expect a chiral center adjacent to ketone carbonyl to be more prone to racemization than a chiral center adjacent to amide carbonyl. The main issue is that we’ve not even synthesized any compounds in which the amide NH of 3-aminopyridines has been replaced with CH2. It is on this basis that I recommend exploring the your idea with BEN-DND-61647d40-20 (or the analogous design derived from EDJ-MED-49816e9b-1) since these are structurally-related to known inhibitors.
My advice to the project management/leadership would be to define targeting the catalytic cysteine as a design theme and then to explore options systematically. I brought the PET-UNK-5ecb6237-1 to the discussion because it’s a ‘nitrile equivalent’ of your design and that of @Ben_DNDi. I also wanted to highlight that exploitation of of the amide NH as a vector for structural elaboration is not as easy as one might think.
@pwkenny our strategy has been very specifically first to generate strong molecular recognition, and only then embark on “covalentising” with very weak electrophilic groups. So no, we do not intend to go back to lower potency compounds: we will start from our front-runners, and consider this as one route to optimisation. (Obviously other strategies might work, but it’s simple: we don’t have the money.)
I’m puzzled that you keep saying the NH is being replaced by CH2; in my design it’s an sp2 carbon.
What reasoning are you applying to think that it would exist in the tautomer “as drawn”? (I didn’t draw it that way - I’m not sure I can control how it’s slurped up.)
PET-UNK-a692de38-1 has many structural alerts - are you concerned? We could only consider sticking stuff on the nitrogen if it’s synthetically simple.
Hi Frank,
Apologies for lack of clarity. I would expect the compound to exist predominantly in the keto tautomer (as depicted/drawn) since there are no structural features that would stabilize the enol form.
My comments about replacing NH with CH2 were in the context of starting points for ‘covalentising’. 3-Aminopyridines have NH between carbonyl and heterocycle. In what I’ll call the ‘benzotriazole’ series the amide is ‘reversed’ and the heterocycle is linked to the carbonyl by CH2. In order to target the catalytic cysteine, I believe that it will be necessary to link (or directly attach) the warhead to the atom that links the carbonyl to the heterocycle. Previous designs such as BEN-DND-61647d40-20 and NIR-WEI-75ed5c39-1 ‘covalentise’ the CH2 linker by forming a carbon-carbon double bond from it. The point that I was trying to make is that starting point for ‘covalentising’ in your FRA-DIA-a1f3a927-1 design is not something that has actually been synthesized (it’s a ketone rather than an amide and it doesn’t even look like any analogs have been made).
With respect to the structural alerts triggered by PET-UNK-a692de38-1, I note that there are examples of approved drugs with the acylhydrazine substructure in their molecular structures. There are also a couple of examples of approved drugs with nitrile bonded to nitrogen. That said, I see the design as a means to getting an idea about potency gains that can be achieved by presenting a nitrile to the catalytic cysteine and there are other possibilities such as PET-UNK-3e354a91-1 that are likely to be more difficult to synthesize. It’s not clear what the structural alerts are based on and there does seem to be some redundancy in the alerts (different alerts hit the same substructure). Furthermore, I would question how competently the substructural targets have been coded (the cyanamide substructure is considered to be an enamine and the hydrazine substructure triggers the ‘oxygen-nitrogen single bond’ alert.
I hope this is clearer and please let me know if you require further clarification.