Topic automatically created for discussing the designs at:
https://covid.postera.ai/covid/submissions/EDJ-MED-0d144977
Now that the discussion link has been put in place, I’ll comment in the more appropriate place.
I would consider imidazole to be a CYP inhibition risk, especially at the periphery of the inhibitor structure. Another issue is that imidazoles tend to be partially protonated under physiological conditions which creates additional possibilities for off-target activity (including efflux). While you need a pendant hydrogen bond donor (or equivalent) for the tetrahydroisoquinoline scaffold to stabilize the bound conformation, I don’t believe this would still be the case for the dihydroisoquinolone scaffold (which lacks the pyramidal neutral nitrogen). Whether or not the hydrogen bond donor does indeed cause efflux, being able to dispense with it increases the range options and gives the design team more room to manoeuvre.
I would strongly recommend synthesizing the ester PET-UNK-c7ac4d9e-1 and/or its spirocyclic analog (which I’ve not actually submitted) since either of these will tell you whether you actually need a pendant hydrogen bond donor. These esters might also tell you whether a pendant amide is a permeability liability and, if the esters are equipotent with the corresponding amides, I’d be looking at 1,3,4-oxadiazole as an ester replacement. Here’s a PROTAC article which reports improvements in permeability resulting from amide to ester replacement. I should stress that I see replacing the pendant amide with an ester as a means for generating information that you can use in design rather than a solution to ADME problems.
@pwkenny : I’ve included the spirocyclic analgoues as JOH-MSK-4bb3d434 for completeness so I can include them in the next batch of free energy calculations.
Thanks, @JohnChodera, and the binding mode predicted for PET-UNK-77d5678a-3 in the PDB file associated with the PET-UNK-77d5678a submission may be helpful. Although my favored replacement for the pendant amide of EDJ-MED-8bb691af-8 is 1,3,4-oxadiazole (non-hydrolyzable, likely to be metabolically stable and less of a CYP liability than imidazole) as in PET-UNK-aa57768f-1, I have suggested the methyl ester to the design team since it is a good way to test whether the amide NH (which carries baggage) is necessary for potency and it should be as easily synthesized as the amides.
It may be worth looking at the effect of ring size with the FEP calculations. Although PET-UNK-c9c1e0d8-4 and PET-UNK-c9c1e0d8-3 are equipotent it is possible that spirocyclic analogs may differ due to increased degree of geometric contraint. My view is that the 6-membered ring would be better for targeting catalytic cysteine (with an electrophilic warhead) or oxyanion hole (with carbonyl oxygen). Hopefully, good preclinical candidates can be identified without having to use these tactics but it’s always good to have some options in case things don’t go according to plan.
@pwkenny . A Me group will surely sterically discourage haem binding so they may very well be CYP-inhib free (test them PK vs the non-Me analogues?)? We had success in this regard in the past. You could also do a EWG screen on the Imid. to modulate pKa? In the right place it could kill 2 birds with one stone, stop CYP inh and reduce protonation of Im?
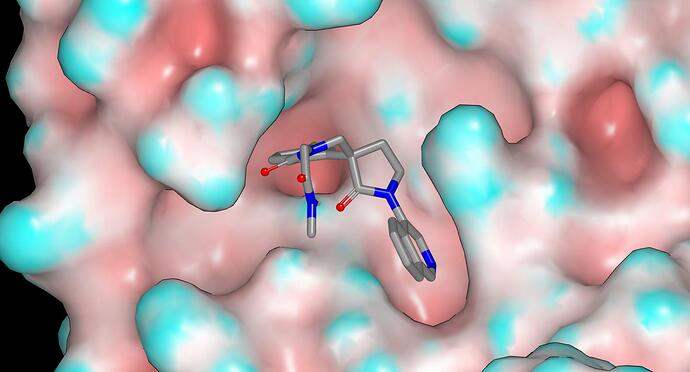
Hi @JSPEN, I’m not suggesting that there definitely will be problems with the imidazoles (or indeed the amides). However, I do think that it would be prudent to think beyond imidazole/pyrazole/triazole if considering heteroaromatic amide replacements since each of these heterocycles has a triply-connected nitrogen that will need to be capped to in order to eliminate the hydrogen bond donor. The pendant amide of EDJ-MED-8bb691af-8 does not appear to form hydrogen bonds with target and binding is likely to incur a penalty for desolvation of the amide NH (there does not appear to be room for a water or a methyl substituent on the amide nitrogen). The PDB file associated with the PET-UNK-14142a25 submission includes predicted binding modes that illustrate the orientation of the pendant amide of EDJ-MED-8bb691af-8 (see PET-UNK-14142a25-1) and I’ve included a graphic below. If there are indeed problems associated with the pendant amide, my view is that synthetic elaboration from the N-methyl of EDJ-MED-8bb691af-8 will be unlikely to solve these them (and may well introduce new problems).
Too many Ns can also be a problem with having high energy content molecules and N2 release? Don;t you just love medchem; you get over one road bump and then hit a mountain of new problems!!!