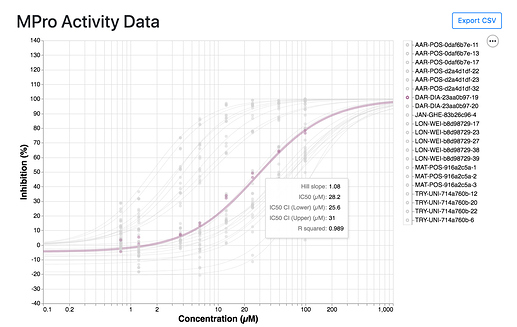
I’ve been looking at the assay results at https://postera.ai/covid/activity_data and have a few questions.
Is there an explanation for the column names in the csv file? Most of these make sense, but I’m not sure what the difference is between ‘IC50 (uM)’ and ‘IC50 (uM) - std’.
What were the criteria for deciding which compounds were tested in dose-response?
Several of the structures are duplicated. They have the same SMILES, but different CIDs
CC(=O)N1CCN(CC(=O)Nc2cccc(S(N)(=O)=O)c2)CC1 JAN-GHE-1d98ec1c-7,LON-WEI-b8d98729-4
CC(C(=O)O)c1ccc2c(c1)[nH]c1ccc(Cl)cc12 AAR-POS-fca48359-19,AAR-POS-fca48359-20,GER-UNI-ec786817-1,MAK-UNK-0d6072ac-4
CC©N©C(=O)C1CCN(C(=O)CCl)CC1 AAR-POS-0daf6b7e-19,VIR-GIT-7b3d3065-3
CNS(=O)(=O)Cc1ccc2[nH]cc(CCN©C)c2c1 ANT-OPE-47f3bb65-1,TOM-OIS-cbf37265-2,LON-WEI-b8d98729-34
COc1ccc2[nH]cc(CCNC©=O)c2c1 ALE-ACE-2610ee4d-1,JOH-MSK-35a8745a-1
CS(=O)(=O)Nc1cccc(C(=O)Nc2cccc(N3CCCC3=O)c2)c1 MAK-UNK-1e8f9e3c-1,MAK-UNK-9159f8ed-6
Cc1ccc©c(S(=O)(=O)N2CCN(C(=O)CCl)CC2)c1 AAR-POS-0daf6b7e-3,WAR-XCH-79d12f6e-6
Cc1ccncc1NC(=O)Nc1cccc(Cl)c1 JAG-UCB-cedd89ab-8,TRY-UNI-714a760b-12
Cc1ccncc1NC(=O)Nc1ccccc1 TRY-UNI-714a760b-4,ALE-HEI-f28a35b5-1
Cc1ccncc1NC(=O)c1cccc(NC(=O)Nc2cccnc2)c1 MAK-UNK-129dcd6f-9,MAK-UNK-129dcd6f-11
N#Cc1ccc(NC(=O)Nc2cccnc2)cc1 JOH-UNK-14e6adc5-2,ANN-UNI-26382800-3
N#Cc1cccc(NC(=O)Nc2cccnc2)c1 JOR-UNI-61e7b1d5-2,ANN-UNI-26382800-4,WAR-XCH-eb7b662f-1,DAR-DIA-23aa0b97-5
O=C(CCl)N1CCC(C(=O)N2CCCCC2)CC1 AAR-POS-0daf6b7e-9,VIR-GIT-7b3d3065-2
O=C(CCl)N1CCN(Cc2cccc(Cl)c2)CC1 AAR-POS-d2a4d1df-35,MED-COV-4280ac29-15,MED-COV-4280ac29-16
O=C(CCl)N1CCN(Cc2cccc3ccccc23)CC1 AAR-POS-d2a4d1df-40,MED-COV-4280ac29-31
O=C(CCl)N1CCN(Cc2cccs2)CC1 AAR-POS-0daf6b7e-17,LON-WEI-8f408cad-5
O=C(CCl)N1CCN(Cc2ccsc2)CC1 MAK-UNK-212f693e-8,AAR-POS-0daf6b7e-13,LON-WEI-8f408cad-7
O=C(CCl)N1CCN(S(=O)(=O)c2c(F)cccc2F)CC1 AAR-POS-d2a4d1df-32,LON-WEI-8f408cad-2
O=C(CCl)N1CCN(S(=O)(=O)c2cccc(F)c2)CC1 AAR-POS-d2a4d1df-23,LON-WEI-8f408cad-4
O=C(CCl)N1CCN(S(=O)(=O)c2cccs2)CC1 AAR-POS-d2a4d1df-22,LON-WEI-8f408cad-3
O=C(CCl)Nc1cccc(N2CCCC2=O)c1 MAK-UNK-7a704a63-10,AAR-POS-0daf6b7e-11,LON-WEI-8f408cad-1
O=C(c1cc(=O)[nH]c2ccccc12)N1CCN(c2ccccc2)CC1 EDJ-MED-78f964c8-1,MAT-POS-916a2c5a-4
Any idea what’s going on here?
Thanks,
Pat