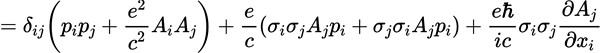“The future is already here – it’s just not evenly distributed" - William Gibson
Sometimes working in Chemistry gives you the eerie feeling that you have seen the future and yet are living in the past. Over many years, humanity has come up with some decent ways to store and pass around data. Putting a table of important supplementary data in a PDF is not one of them. Turns out that pictures are not a great way of making searchable databases either.
Each time I have to redraw a molecule from a paper to search it, or reproduce a route from a patent in Chemdraw, I’m reminded that this is all because we’ve done this to ourselves. But while I expect changing these norms will be slow, one of my favorite tools of the last five years is luckily coming to Chemistry to help ease the pain while we continue to drive ourselves mad.
The tool is Mathpix. I encourage you to check out their website to learn more, but here’s the problem they originally solved for me: You are reading a math paper and a really complicated equation comes up. You want to put that equation in your notes, but typing math is not fun. So instead of typing it again, just take a screenshot with MathPix, and the formula is written for you! This visual on their site is likely a more helpful explanation:
For those who are wondering why this was a big problem, typing math in LaTeX can feel a bit like drawing complicated natural products in ChemDraw, but you also need to know a whole language of commands. For example, here’s a random equation I took from the internet.
Mathpix told me that this corresponds to the LaTeX command
$=\delta_{i j}\left(p_{i} p_{j}+\frac{e^{2}}{c^{2}} A_{i} A_{j}\right)+\frac{e}{c}\left(\sigma_{i} \sigma_{j} A_{j} p_{i}+\sigma_{j} \sigma_{i} A_{j} p_{i}\right)+\frac{e \hbar}{i c} \sigma_{i} \sigma_{j} \frac{\partial A_{j}}{\partial x_{i}}$
Now imagine writing pages of that; it can get quite tedious.
Isn’t all of Chem(pix) just applied Math(pix)?
Anyway, you can already see the parallels. Why can’t we as chemists have a tool where we screenshot images of molecules in papers or patents and instantly get the SMILES to paste into other tools? Well according to their email today, we are quite close!
Copying directly from their email:
Beta chemical diagram recognition & ChemDraw compatibility
We have launched a beta of image to SMILES conversion for recognition of chemical diagrams. This is a very preliminary version of the recognition but we were too excited not to share! Accuracy is still a big work in progress. More improvements, including handwritten chemical diagram recognition, coming soon!
Now let me demonstrate how slow I am at drawing…
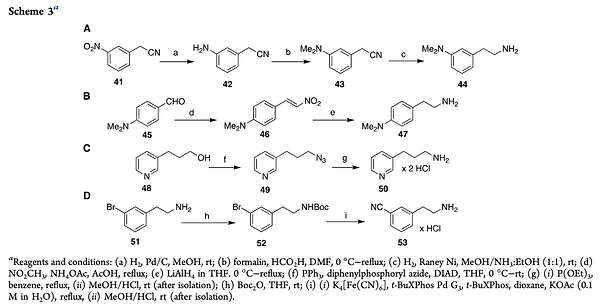
Let me give an example from a paper I came across the other week. I don’t want to name names, or journals because it’s not productive (plus it’s a systemic issue, not one paper). Anyway, basically I wanted to check if the building blocks 44, 47, 50, and 53 in the image could be bought directly without making them from scratch (mostly for curiosity’s sake).
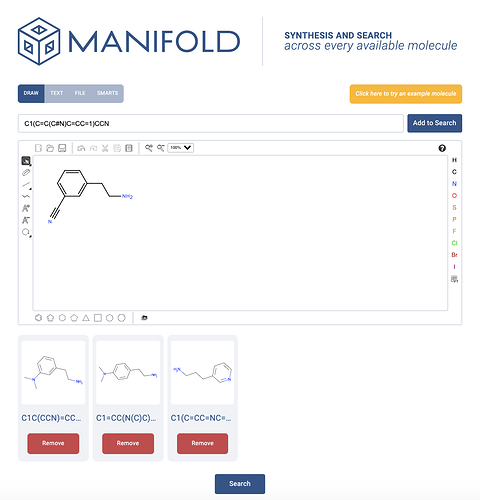
This paper was better than many, and did have the SMILES strings for all intermediates and targets in the SI (supplementary information). However, I had downloaded the paper and just wanted them quickly without going back to download the SI. So I ended up just drawing the molecules in Manifold.
Try it yourself on Manifold (anyone can make an account). It took me about 1 minute 15 seconds (not racing), but then I realized I forgot a methylene linker in the molecule (50). So probably about 2 minutes overall. Some of you will finish much quicker and swiftly understand why I am so happy for a new tool to help me. Hopefully at least one of you empathizes.
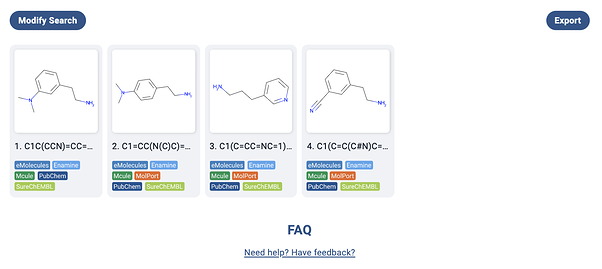
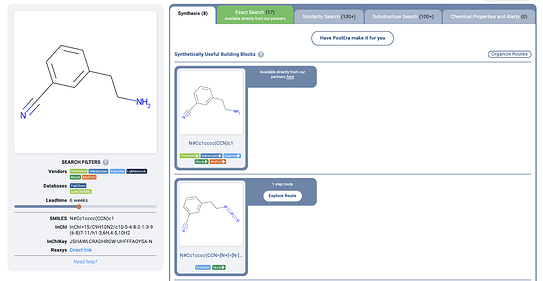
On the plus side, when I pressed search, I found that all the building blocks were in stock from popular vendors – so at least I could save a lot of time and money on synthesis without having to go through 3-4 step routes!
If none of you chemists see the issue, imagine being a biologist trying to send a colleague some chemical probes from past papers that would be interesting to test. Drawing compounds with a somewhat rusty knowledge of organic chemistry is a bit like attempting to learn “Happy Birthday to You” on the piano without being able to read music or knowing where to place your fingers. It can be done, but it’s a lot harder than for someone who can do those things. (Also, one day try explaining to a non-chemist where the bonds should go when drawing 5-membered aromatic heterocycles).
The progress so far
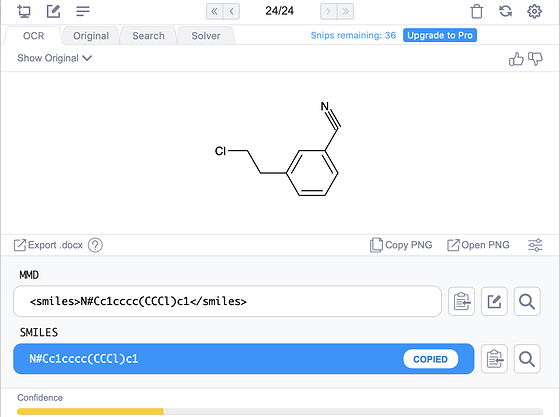
Now for Mathpix, it’s still in beta, so I won’t pass too much judgement. From their math products, I know they care about correctness and speed and ease of use, so I expect great stuff. If you are interested in how it does for the aforementioned four molecules, these are particularly hard because of the presence of salts and abbreviations such as “NMe2”.
On the last molecule, here is what my screenshot of the original paper produces (the bend in the nitrile is an interesting artifact of their recognition):
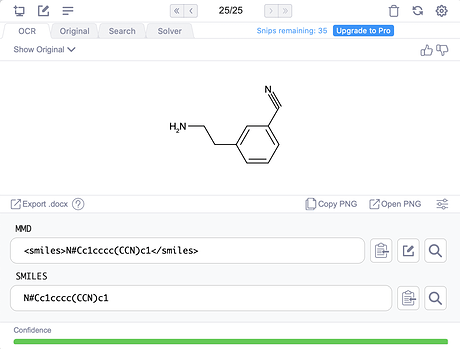
It’s close, but unfortunately the salt HCl salt messes up recognition of the amine. Obviously, these things should get better. At least the confidence shows the model knows what it doesn’t know! From other experiments, spirocycles, bridgehead atoms, and abbreviations like N3 for azide can cause issues in molecules. Going back to this molecule, if we try Mathpix on the cleaner molecule image on Manifold (where we use the wonderful RDKit to draw images), then all is well:
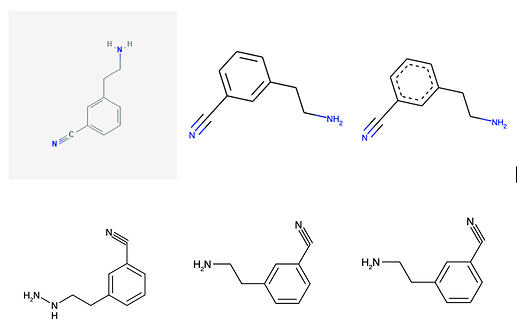
As a quick experiment, let me write some code to get images from other sources and see if it gets all of them right. I assume this type of data collection/augmentation is done during training of the algorithm (along with rotations, distortions, etc…). Anyway, here’s a quick code snippet to get some other images of the molecule:
import pubchempy as pcp
from rdkit import Chem
from rdkit.Chem import AllChem
c = pcp.get_compounds(smiles, namespace='smiles')[0]
pubchem_id = c.cid
pcp.download('PNG', 'pubchem_test_mol.png', pubchem_id, 'cid', image_size='300x300')
Chem.Draw.MolToFile(Chem.MolFromSmiles(smiles), 'rdkit_fuzzy_test_mol.png', kekulize=False)
Chem.Draw.MolToFile(Chem.MolFromSmiles(smiles), 'rdkit_fuzzy_kekule_test_mol.png', kekulize=True)
As you can see, with the original images at the top, and recognized mol at the bottom, only the pubchem depiction currently gives it some trouble.
Again, this is only the beta version, and I assume Mathpix is working hard to fix these issues. I’m guessing it will be a bit before we get a perfect SMILES translation of the drughunter.com small molecule of the year or Taxol but from knowing how much time their math tool saved me, I am very excited for what’s to come.
Some other background
This certainly is not the first optical chemical structure representation (OCSR). In fact, there is a nice recent JCIM article going through the history of these tools A review of optical chemical structure recognition tools | Journal of Cheminformatics | Full Text that date back to the early 90s. As far as I can tell, most of these programs focus on going quickly through large batches of documents and extracting structures and other chemical information to feed into databases. The paper also has some nice benchmarks of speed and accuracy on datasets much larger than my very limited example.
However, I imagine Mathpix’s tool (can I call it Chempix?) will cater to a different user base, given their great focus on user interface and integration with other tools. The tool will be useful for students who want to digitize their handwritten notes, or readers of journals who want to query molecules they are reading about in papers.
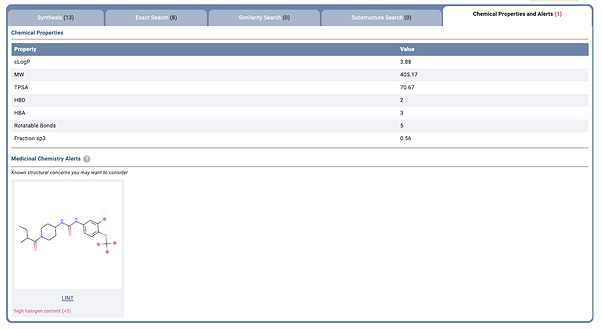
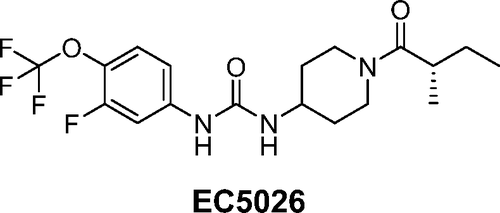
For example, I went to the current issue of J Med. Chem and used the tool on the graphical abstract of Movement to the Clinic of Soluble Epoxide Hydrolase Inhibitor EC5026 as an Analgesic for Neuropathic Pain and for Use as a Nonaddictive Opioid Alternative, Mathpix got it right (at least the racemate form CCC(C)C(=O)N1CCC(NC(=O)Nc2ccc(OC(F)(F)F)c(F)c2)CC1) in under a second, and 2 seconds later I had the physical properties up in Manifold. That’s a pretty nice companion to have when reading a paper without lots of drawing.
Overall, I hope this tool keeps being rapidly developed and I’ll surely be a user. As someone who hates going between papers and drawing tools, while trying to remember if it’s the urea or the amide, the pyrazole or the triazole, the indole or the purine, this will definitely be welcomed. I actually told someone to develop this a year ago, but I’m glad Mathpix decided to expand to Chemistry!
Next, I want a predictive keyboard for typing SMILES, then I can do away with drawing all-together. And then I’ll never have to teach anyone the conventions of IUPAC naming, and we can all just communicate in SMILES, and the first week of undergraduate orgo won’t have to be classes on naming conventions, and I’ll never have to remember what a tetrahydroquinoline or phthalazine is again… And I’m kidding of course. But only slightly. Let’s stop putting tables of data in PDFs before we solve that issue.
-Matt