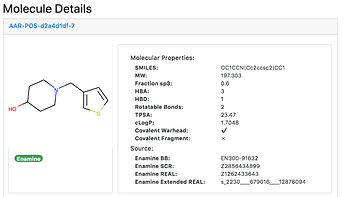
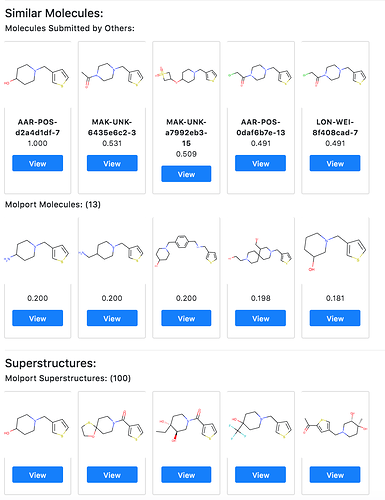
As far as we can tell, the bolus of Moonshot submissions were not generated by algorithm, but by people’s eyeballs. There must therefore be a big opportunity to trawl algorithmically the very large modern databases and enumerations of synthetically accessible compounds (e.g. Enamine REAL), for things that magically merge several or many of the observed fragments and/or their interactions.
Of course, the rapid scoring methods introduced in the other topic in this category are probably key to doing this.
We are therefore inviting discussions/suggestions of how to do this. Also welcome would be actual enumerations: these will be discussed and considered for actual compound purchase, if they are sufficiently convincing and they are available before we run out of time and budget.