Given recent interest in the tetrahydoisoquinoline-based scaffold (in particular, the methylsulfonyl analog MAT-POS-dd3ad2b5-4), it may be helpful to share some observations on protein structures for chromane-based ligands. The IC50 for MAT-POS-dd3ad2b5-4 does not to appear to have been uploaded (at least the last time I checked) and I’m assuming, based on notes associated with the FRA-DIA-6238d354 submission, that it is about 100 nM. I’ll mention @alphalee @frankvondelft @JohnChodera @londonir @mc-robinson @edgriffen in case these observations are of interest.
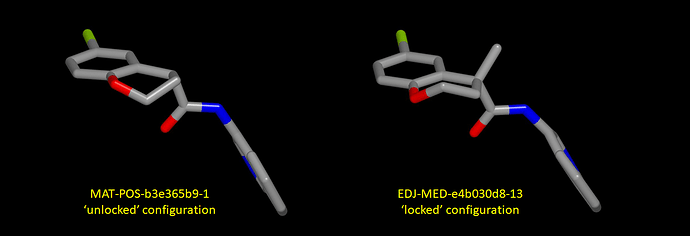
The puckering of the chromane ring appears to differ according to whether or not the configuration of the chiral center has been ‘locked’ by quaternization. This can be seen in a graphic showing the crystallographic ligands from X11612 (crystallographic ligand: MAT-POS-b3e365b9-1 ; ‘unlocked’ configuration) and X12207 (crystallographic ligand: EDJ-MED-e4b030d8-13; configuration ‘locked’ by methyl substituent).
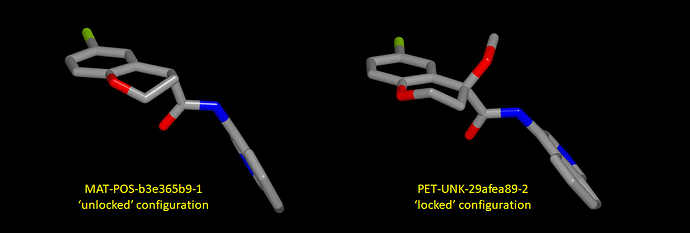
Here’s a graphic showing an analogous difference and in this case the ligand for which the configuration is ‘locked’ by the methoxy substituent (PET-UNK-29afea89-2) is from the P0157 structure).
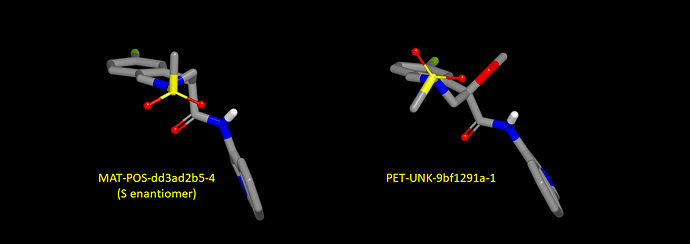
Sulfonamide nitrogen is typically pyramidal and the C-S tends to be anti with respect to the nitrogen ‘lone pair’. Here’s a graphic showing predicted bound conformations for the S enantiomer of MAT-POS-dd3ad2b5-4 (‘unlocked’ configuration) and PET-UNK-9bf1291a-1 (‘locked’ configuration). ‘Locking’ the configuration makes the S-C vector point in the opposite direction and this has implications for design.
1 Like
Really interesting Pete, do you have any insights into the energy difference between the two sulfonamide conformations? We have the methoxy tetrahydroisoquinoline sulfonamide in progress for synthesis, though it is non-trivial.
Hi Ed,
I should have pointed out that the sulfonamide models were generated by allowing each tetrahydroisoquinoline to ‘inherit’ the puckering of the chromane from which it was derived and I don’t have any real insights at this point. I think that the puckering of the tetrahydroisoquinoline will be a more of an issue for sulfonamide library design (with a view to increasing potency) than if tetrahydroisoquinoline sulfonamides are seen primarily as a means to improve ADMET characteristics. My view from examining proposed binding modes for the methylsulfonyl and acetyl tetrahydroisoquinolines in the context of the protein structure is that structural elaboration from the tetrahydroisoquinoline nitrogen would be unlikely to result in useful potency gains. The pdb files associated with the PET-UNK-9bf1291a (methoxy-locked) and PET-UNK-162c14b2 (unlocked) submissions include proposed binding modes for both methylsulfonyl and acetyl tetrahydroisoquinolines.
One question is whether the project leadership would be prepared to nominate a candidate with an ‘unlocked’ chiral center next to a carbonyl group (I’m not sure how this would be viewed by regulators in the light of thalidomide). I would be nervous about an ‘unlocked’ chiral center and, given that the configuration can be locked with methyl (EDJ-MED-e4b030d8-13) or methoxy (PET-UNK-29afea89-2) without compromising potency, it would seem perverse proceed with an ‘unlocked’ chiral center.