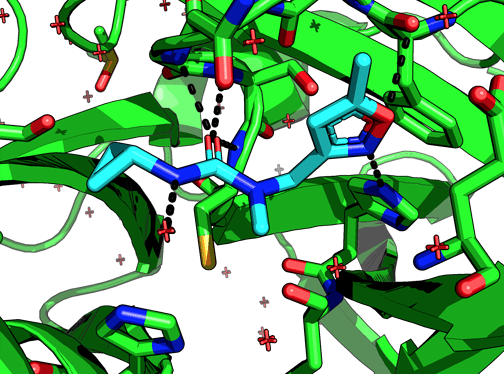
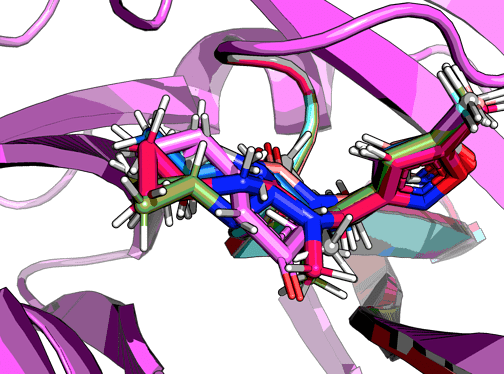Among the non-covalent fragments this one looks unique in that the cPr accesses a pocket (which looks quite large) that no other fragment does. Does the NMe bias the urea conformation to direct the cPr to this pocket? Or, to put it another way, if we remove the NMe would we lose potency?
The N-methyl group is solvent facing in x0397, so the idea is that unmethylated it would be able to hydrogen bond with the protein, say N142.
On the Diamond website one can find the list of compounds in the libraries that failed. That would most likely be the most informative place to look.
However, in silico I quickly tried the following (placed with my algorithm Fragmenstein and docked them with pyrosetta’s DockMCMProtocol mover:
-
x0397:
Cc1cc(CN(C)C(=O)NC2CC2)no1: control behaved -
unmethylated:
Cc1cc(CNC(=O)NC2CC2)no1: nothing happened -
diacetyl:
Cc1cc(CC(=O)C(=O)NC2CC2)no1: nothing happened -
alkane:
Cc1cc(CCC(=O)NC2CC2)no1: nothing happened
Changing the keto group gave funkier results:
-
keto2alkene:
Cc1cc(CN(C)C(=C)NC2CC2)no1: pushed away by 1.3Å on the axis of the sidegroup -
keto2methylamine:
Cc1cc(CN(C)N(-C)NC2CC2)no1: 0.5Å RMSD -
keto2ethylamine:
Cc1cc(CN(C)N(-CC)NC2CC2)no1: 0.6Å RMSD -
keto2butylamine:
Cc1cc(CN(C)N(-CCCC)NC2CC2)no1: escapes into solution
For backbone stapling (for fun) I tried:
-
quinone:
O=C1C(Cc2noc(C)c2)=CC(=O)C=C1(C3CC3)
So nothing much happens. From the looks of it the cyclopropane be accomodated in different torsion angles.
Nice work, Matteo, thanks!
Truth be told, x0397 is one of the compounds in the 4-compound merger by Frank Von Delft —Submission FRA-DIA-8640f307 —so I really wanted to try some rigid backbone (hence the rando quinone), but they did not work on the latter.