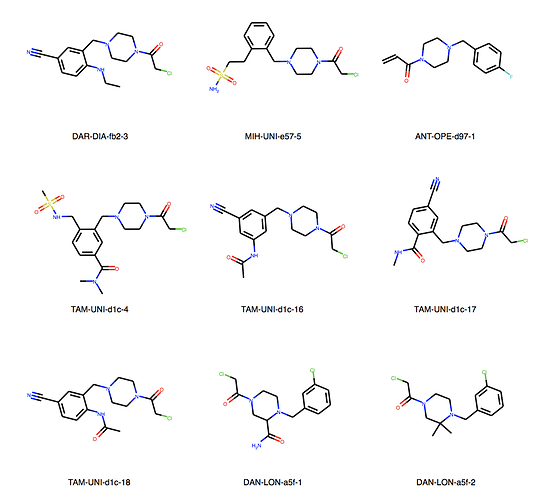
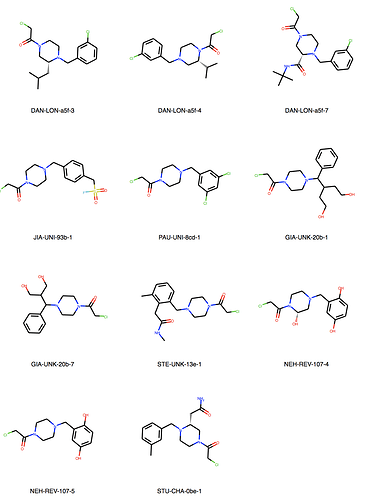
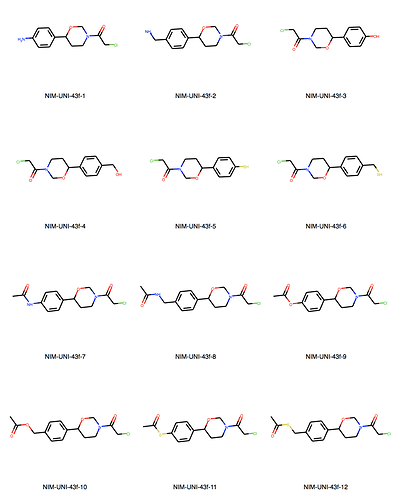
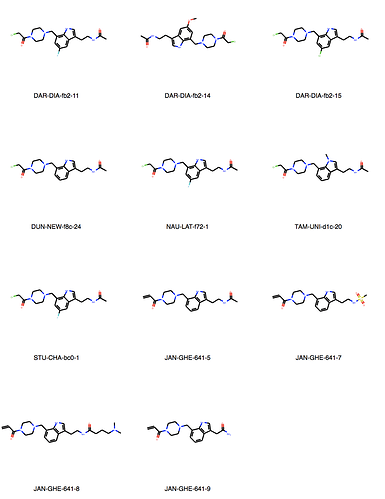
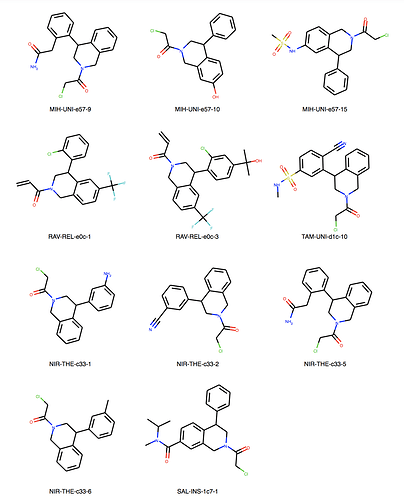
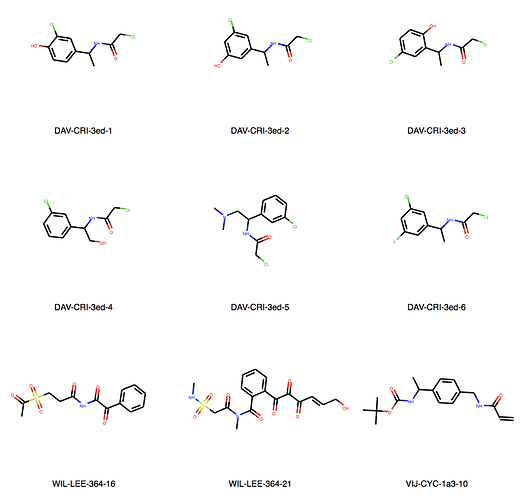
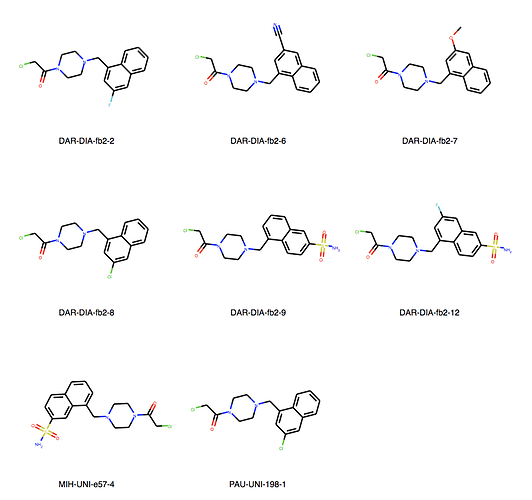
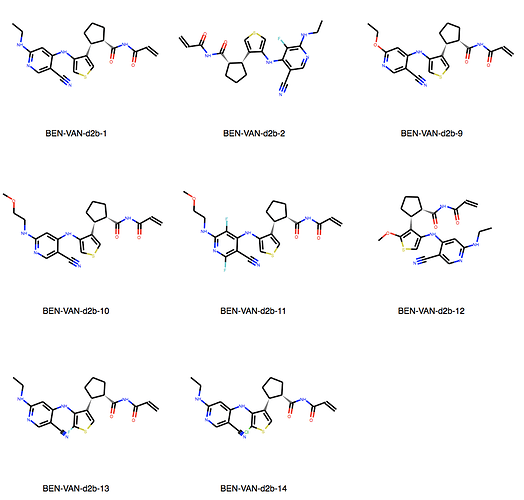
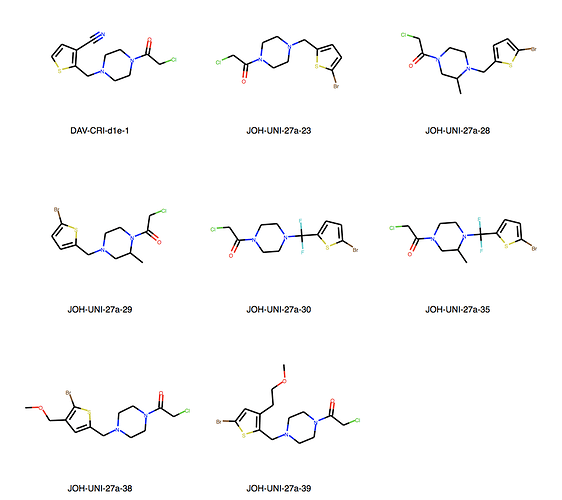
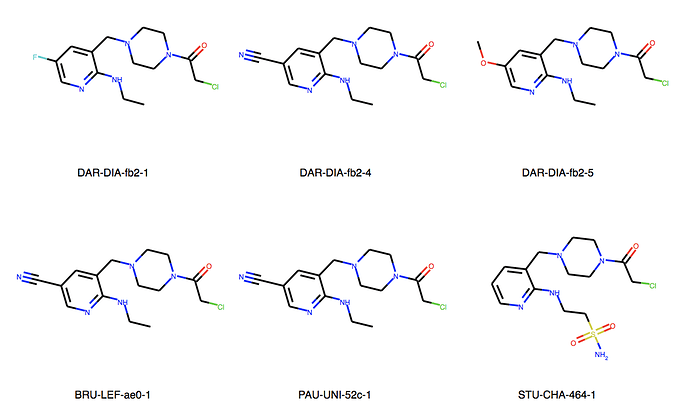
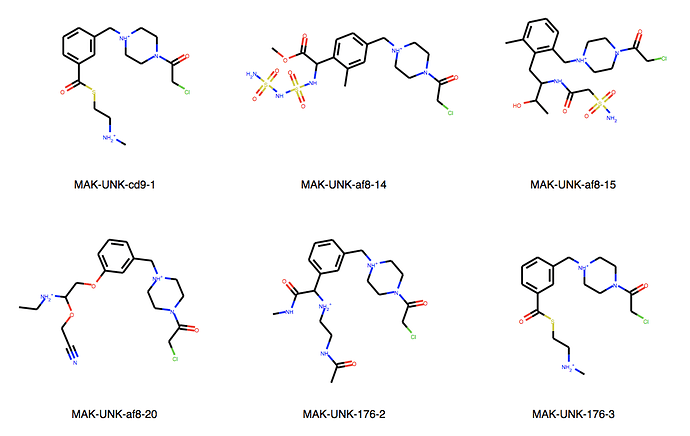
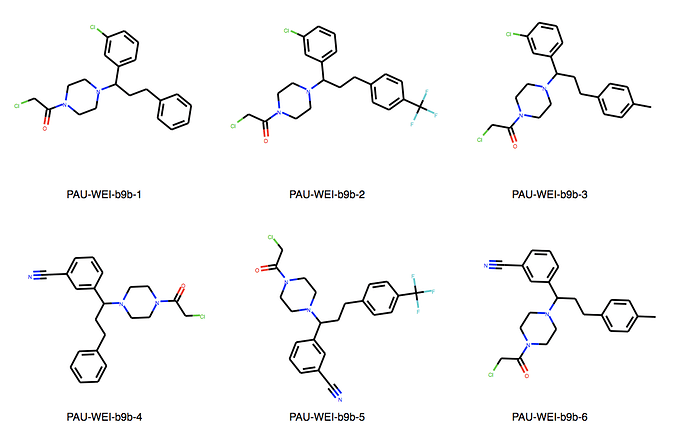
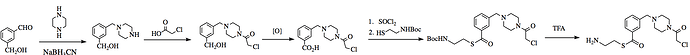We can explore the cluster in greater detail. Cluster 1:
Hi Alpha,
Of course checking if my own entries are in there. We’re all human, aren’t we? But found none. I would say JOO-PER-58e would at least fit in cluster 6. And there are more. Are they all singletons or is there something amiss? They were designed to be all covalent …
best Joost
Ah, the way the Murcko scaffold is defined in rdkit is exact atom match. Cluster 6 contains naphthalenes whereas your design is a isoquinoline. I’ll think about defining the scaffold more generally.
Yes, there are a lot more singletons. I’ve just listed clusters with more than 5 molecules.
Thanks for that! Appreciate your efforts, fighting the virus on saturday morning
Data warrior has a couple options for Murcko scaffolds. Murcko scaffold, Murcko skeleton and most central ring system. It would be trivial to merge these into the master file. Let me know if you want to go this route, and if so will you do it or do you want me to generate the columns.
About cluster 10. They are all mine, designed in SeeSAR Inspirator. Note that the first and last are exactly the same. It probably happened because they were written differently:
- MAK-UNK-cd9-1: N+©([H])C(C(SC(C1C=C(CN+2CC(N(CC2([H])[H])C(CCl)=O)[H])C=CC=1[H])=O)[H])[H]
- MAK-UNK-176-3: NH2+C
So it’s exactly 5 molecules, not more, sorry about that.
I tried to see how complex the synthesis would be on https://app.molecule.one/, and it can’t finish.
Hopefully PostEra/Enamine will have more luck.
HI,
is there somewhere we can pick up the latest list of the covalent suggestions with the attachment points or the covalent functional groups?
Thanks, Bobby
Hi @RGlen, The closest I have is https://raw.githubusercontent.com/mc-robinson/COVID_moonshot_submissions/master/covid_submissions_all_info.csv , which lists if the compound contains a “covalent warhead” (and specifies acrylamide, acrylamide_adduct, chloroacetamide, chloroacetamide_adduct, vinylsulfonamide, vinylsulfonamide_adduct ). However, I believe @londonir has the specific file you are looking for.
@AnthonyA, am I correct in this?