May '20
May '20
▶ frankvondelft
@matteoferla, how does Fragmenstein score these guys? Particular questions:
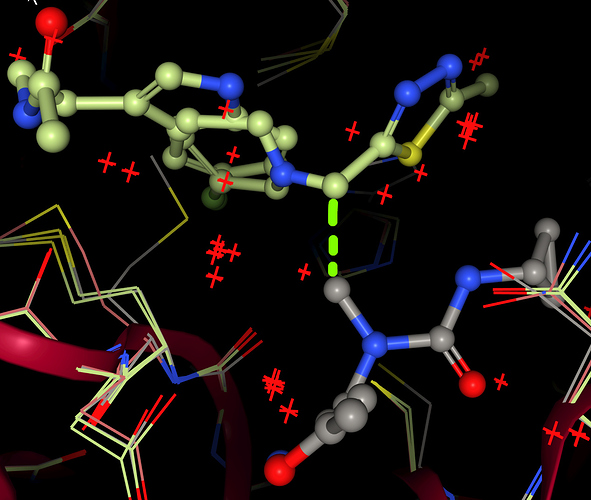
- The atoms are meant to map directly from hit to design - does your algorithm figure it out?
- The protein conformation down in H61/M165/C145 is quite varied for the different inspirations - if you aren’t already, then you need your heuristic to spot which sidechains move, and include them in the things that move and are tracked. (I think…
 )
)
May '20
May '20
May '20
▶ frankvondelft
Sorry, I did not notice there were two submissions.
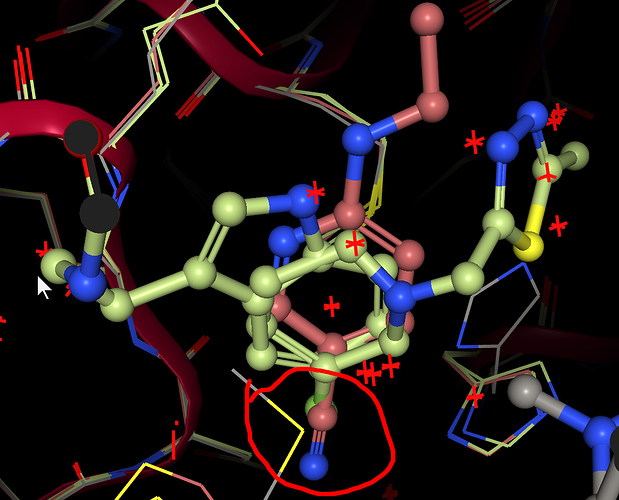
Okay. I am sorry to say, but your SMILES are wrong. Your heterocycles don’t come out planar. These are aromatic, because you did not specify the Hydrogen or positive charge.
CC(=O)NCCc1c2cc(F)cc(C(CN(Cc3cc(C)on3)C(=O)NC3CC3)c3nnc(C)s3)c2[nH]c1CC(=O)NCCc1c2cc(C#N)cc(C(CN(Cc3cc(C)on3)C(=O)NC3CC3)c3nnc(C)s3)c2[nH]c1CC(=O)NCCc1c2cc(Cl)cc(C(CN(Cc3cc(C)on3)C(=O)NC3CC3)c3nnc(C)s3)c2[nH]c1
Results
|
name |
mode |
∆∆G |
comRMSD |
N_constrained_atoms |
runtime |
disregarded |
| 0 |
FRA-DIA-8640f307-1 |
none |
-15.6921 |
0.88076 |
38 |
16.8203 |
[‘x0305’] |
| 1 |
FRA-DIA-8640f307-2 |
none |
-13.4534 |
0.886753 |
37 |
16.4181 |
[‘x0305’] |
| 2 |
FRA-DIA-8640f307-3 |
none |
-13.8871 |
0.889806 |
37 |
16.9554 |
[‘x0305’] |
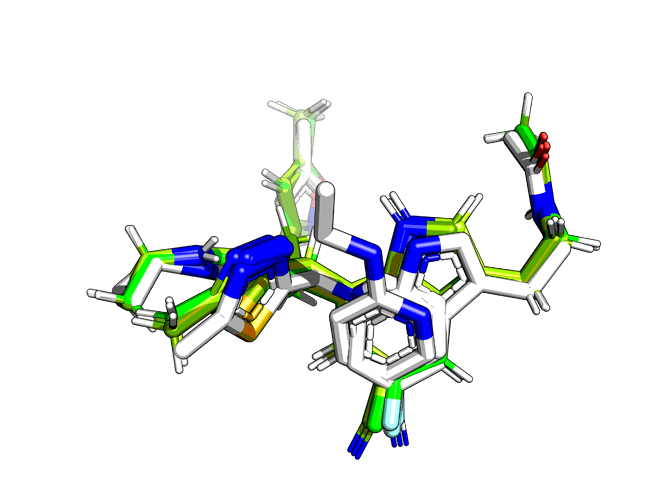
These are minimised, so they work fine:
2 replies
Jun '20
▶ matteoferla
Jun '20
▶ matteoferla
@AnthonyA, I hadn’t seen until now those minimisations that @matteoferla had run; according to that, these are very happy compounds.
Please let’s discuss how to get them made…
Jun '20
▶ frankvondelft
Hi @frankvondelft, @matteoferla, sorry I didn’t see this earlier. This should now be fixed. Just a bit of the reason for why users cannot edit their SMILES directly:
Implementing this is a pain, because once a compound is ordered/made/assayed, we don’t want the identity to change in our system from what was actually ordered/made/assayed. Furthermore, even before that, CDD integration messes up due to the SMILES being different.
I also believe @alphalee said he has routes for these if you want to get them made.
 )
)